
Primary diffuse large B-cell lymphoma of the central nervous system: molecular and biological features of neoplastic cells
Introduction
Central nervous system diffuse large B-cell lymphoma (CNS-DLBCL), formerly named primary CNS lymphoma (PCNSL), is an extranodal aggressive lymphoma manifesting solely in the brain, spinal cord, leptomeninges or in the eye and represents the vast majority of CNS-lymphoma cases (1,2). Since other articles published on the same special issue describe the epidemiology, the pathology, the clinical and therapeutic approaches for CNS-DLBCL, as well as of secondary central nervous system involvement, here we will focus on the genetics and the biology of CNS-DLBCL cells. Remarkably, the main patho-mechanisms for CNS-DLBCL had already been reported before 2012 and were subsequently confirmed by numerus publications from different groups (Figure 1). Epstein-Barr virus positive CNS-DLBCL show a totally different pathogenesis (3), and we will omit this type of CNS-lymphoma from this review article.
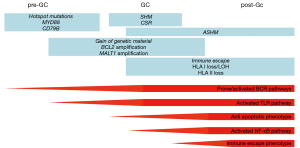
CNS-DLBCL express non-switched self-reactive somatically mutated immunoglobulins
Tumor cells of CNS-DLBCL present immunoglobulin (Ig) heavy and light chain genes with somatic mutations, compatible with their origin from mature B-cells that have encountered antigen and have undergone T-cell dependent somatic maturation (4-12). IGHV4 family genes are frequently rearranged, with a preference for the IGHV4-34 gene segment (4,5,7-9,11).
The tumor cells do not show a successful class switch, as such, they express neither IgA, nor IgE, nor IgG, but only IgM and IgD (13).
The immunoglobulins expressed by CNS-DLBCL cells do not react against self-antigens commonly involved in autoimmune disorders, but they do bind self-proteins, including proteins expressed in the central nervous system, such as GRINL1A, ADAP2 and BAIAP2 (9), or N-hyperglycosylated SAMD14 and neurabin-I (11).
In agreement with the mutated immunoglobulin heavy chain genes, CNS-DLBCL cells also present aberrant somatic hypermutation (ASHM) that affect a series of genes, such as BTG2, HIST1H1E, KLHL14, MYC, PAX5, PIM1, RHOH, or SUSD2 (10,14-16).
CNS-DLBCL have an activated B-cell like (ABC) DLBCL phenotype
DLBCL represents an heterogenous group of diseases (17-20). Studies performed in systemic DLBCL have first identified two main subgroups, the germinal center B-cell like (GCB) and the ABC DLBCL, based on their expression profiles suggestive of their potential cell of origin (COO) (21-24). GCB and ABC DLBCL have differences in genetics and biologic features and, importantly, in the clinical outcome when patients are treated with the standard R-CHOP treatment (18,24). The vast majority of CNS-DLBCL are constituted by non-GCB DLBCL. This has been reproducibly demonstrated in many studies, largely performed using immunohistochemistry (15,25-42), but also via gene expression profiling (33,43-46).
Differently from what observed in systemic DLBCL, the prognostic impact of the COO in CNS-DLBCL seems to be minimal, with the vast majority of studies not finding any difference in the clinical outcome between patients with ABC or GCB CNS-DLBCL (31-43). Fukumura et al. reported an inferior progression-free survival (PFS) for non-GCB than GCB cases in a series of 39 patients (15). Shi et al. reported a borderline inferior 3-year overall survival (OS) for ABC than GCB-PCNS DLBCL when considering 77 patients treated half with methotrexate-based scheme and half not, but the difference was not maintained when they analyzed the outcome based on treatment modality (29).
CNS-DLBCL cells belong to a specific ABC DLBCL cluster
In the last few years, ABC and GCB DLBCL have been further subclassified in additional sub-groups that share more homogenous genetic lesions (47-50). In particular, a specific ABC DLBCL subset has been recurrently identified, although named in different ways: C5 (47), MCD (48,49) or MYD88 cluster (50). These lymphomas are characterized by the presence of genetic events that activate the B-cell receptor (BCR), the Toll-like receptor (TLR) and NF-κB signaling (often concomitant CD79B mutations and MYD88 L265P mutation; PIM1 mutations), block the terminal B-cell differentiations (TBL1XR1 and/or PRDM1 mutations), deregulate the cell cycle (CDKN2A/B deletions), allow the immune escape (mutations or deletions of HLA-A, HLA-B, HLA-C and CD58), or protect from apoptosis (BCL2 gains), plus additional mutations in KLHL14 or ETV6 (47-50).
Importantly, CNS-DLBCL cells exactly presents a pattern of lesions overlapping with this newly described MCD subtype. Whereas CNS-DLBCL, or more general CNS-lymphoma, refers to a lymphoma manifestation in the CNS, the term CNS-DLBCL should be restricted for CNS-DLBCL of this MCD subtype. Different studies have reported a very high frequency of mutations occurring in CD79B, MYD88 (10,15,16,30,33,51-73), PIM1 (10,15,30,33,53,56-60,65,71,72) and TBL1XR (56,57,59,67,72), BCL2 locus gains (25,39,40,58,74,75), inactivation of CDKN2A (30,55,58,59,62,69,76-87), PRDM1 (59,75,81,85-92) and ETV6 (57,58,93), and genetic lesions leading to immune evasion (15,56,59,78,81,85-87,94).
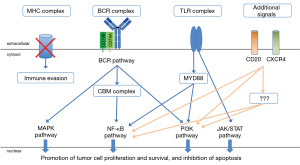
The BCR/TLR/NF-κB signaling is active in CNS-DLBCL
Multiple mechanisms sustain the constitutive activation of the BCR/TLR/NF-κB signaling. As in the C5/MCD/MYD88 cluster, mutation of CD79B and MYD88 usually co-occur. The MYD88 mutations are mostly represented by the c.794T>C substitution resulting in leucine 265 replaced by proline (L265P). Hotspots are also observed in CD79B, with replacement of tyrosine 196 with either asparagine (Y196N), aspartic acid (Y196D), cysteine (Y196C), histidine (Y196H) or serine (Y196S) as the most observed. A lower frequency of MYD88 L265P mutations has been reported in PCSNL from Hispanic versus non-Hispanic patients: 27% (5/18) vs. 66% (8/12) (53).
The gene coding for the PIM1 serine/threonine kinase is among the most mutated. PIM1 is known to be involved in lymphomagenesis (95-98). BCL6 transgenic mouse models have shown that PIM1 cooperates with BCL6, recurrently translocated in CNS-DLBCL (99,100), to give lymphomas (97). Pharmacological and genetic preclinical experiments in cell lines derived from ABC DLBCL, primary mediastinal large B-cell lymphoma and from Hodgkin lymphoma indicate a role for PIM1 in sustaining the lymphoma cells survival, activating NF-κB and JAK STAT signaling and immune evasion (98,101,102).
Similarly to systemic ABC DLBCL, recurrent gains are observed at 3q12.3 leading to the overexpression of NFKBIZ gene coding for the NF-κB co-activator IκB-ζ (58). Functional experiments performed on systemic DLBCL cell lines show that NFKBIZ silencing induces apoptosis and reduces cell proliferation and expression levels of IκBα and BCLXL (58). A similar role might be envisioned also for the NF-κB co-activators IκB-α and IκB-ε, coded by NFKBIA gene and NFKBIE, also recurrently mutated in DLBCL (103,104).
Kaulen et al. have reported mutations in SLIT2 in 3/6 CNS-DLBCL studied by whole exome sequencing (72). Mutational pattern and functional experiments performed, in the non-lymphoma model HEL293T cells transfected with mutated or wild type SLIT2, suggest that the gene might have a tumor suppressor function, negatively regulating WNT- and NF-κB signaling (72). Its inactivation would contribute to NF-κB signaling in CNS-DLBCL.
TERT (telomerase reverse transcriptase) mutations have also been reported in 16% (16/49) of CNS-DLBCL (105). TERT is involved in the maintenance of telomeres and it is both an NF-κB target and NF-κB cofactor. Thus, the mutations would represent an additional mechanism to active BCR/NF-κB pathway (105).
PRDM1 gene, coding for the plasma cell master regulator BLIMP1, is mapped at 6q21, a region commonly deleted in CNS-DLBCL, with DNA losses that often encompass also TNFAIP3, coding for A20, a negative regulator of NF-κB target, and other potential tumor suppressor genes (59,75,81,85-92,106). A similar pattern with a large genomic aberration potentially deregulating both B-cell terminal differentiation and NF-κB, is also observed in systemic ABC DLBCL.
BAFF (B-cell activating factor of the tumor necrosis factor family) and APRIL (a proliferating inducing ligand) are among the signals that lead to activation of the NF-κB pathway in mature B-cells. In particular, APRIL binds to two receptors, TACI (transmembrane activator and CAML interactor) and BCMA (B-cell maturation antigen), while BAFF binds TACI, BCMA, and also BAFF-R (BAFF-receptor).
CNS-DLBCL cells express all three receptors (107,108). The production of the ligands by astrocytes, as shown for BAFF, or, possibly by the neoplastic cells themselves, would contribute to the survival of tumor cells (107-109). Also, the soluble forms of BCMA and TACI (110,111) have been reported elevated in the cerebrospinal fluid (CSF), but not in the serum of CNS-DLBCL patients (110). Similarly, both APRIL (112) and BAFF (111) can be detected in the CSF of CNS-DLBCL patients.
Muta et al. have suggested that another mechanism that might contribute to NF-κB activation in CNS-DLBCL cells is the ghrelin/GHS-R (growth hormone secretagogue receptor) axis (113). CNS-DLBCL cells express both proteins (113) and, in glioma cells, ghrelin activates NF-κB signaling and induces migration of the cells (114).
Finally, the activation of the NF-κB and JAK/STAT signaling, characteristic of the ABC phenotype, are reflected by the presence of secreted factors such as interleukin 10 (IL-10) and of CXC chemokine ligand 13 (CXCL13), which can be detected in CSF, representing a potential diagnostic biomarker and tool to follow patients during therapy (115-130).
CDKN2A is frequently inactivated in CNS-DLBCL
CDKN2A inactivation is very common in CNS-DLBCL (30,55,58,59,62,69,76-87,131), again reflecting its similarities with systemic ABC DLBCL C5/MCD/MYD88 cluster (47-50). Braggio et al. has reported over 80% of cases with CDKN2A inactivation by mutations and/or deletions at 9p21.3 (56). Nayyar et al. showed biallelic loss in 44% of 6 CNS-DLBCL cases (69). Cobbers et al., reported homozygous deletion in 8/20, plus heterozygous loss in additional two cases (77). A mechanism of CDKN2A inactivation is also mediated by promoter hypermethylation (77,131). It is worth of mentioning that CDKN2A losses are matched with a high level of genomic complexity, including deletions at 3p14.2 affecting the tumor suppressor FHIT (58).
TBL1XR1 mutations and reprogramming the differentiation
Mutations in TBL1XR1 were first described in CNS-DLBCL by Gonzalez-Aguilar et al. (59), and were considered to play a regulatory role in the NF-kB pathway. The underling mechanism is much more complex, because TBL1XR1 mutations will impair plasma cell differentiation. Thereby shifting the B-cell differentiation to non-class-switched IgM memory B-cells with an aberrant cyclic reentry to new germinal centers (132,133), which reflects the phenotype of the tumor cells in CNS-DLBCL (13,44).
Epigenetic regulators are only marginally affected
Mutations in genes coding for epigenetic regulators, in particular EZH2, CREBBP, EP300, are very uncommonly mutated in CNS-DLBCL (15,30,34,57,72,134), and only KMT2D seems to be mutated in 10-40% of the cases (15,30,33,71). The pattern is in agreement with what reported in systemic ABC DLBCL belonging to the C5/MCD/MYD88 cluster (47-50).
TP53 can be mutated in CNS-DLBCL
TP53 mutations have been reported at very different frequencies across studies (15,26,33,71,76,77,82,85,90). Perhaps reflecting the ABC DLBCL C5/MCD/MYD88 cluster (47-50), CNS-DLBCL shows a generally lower frequency of TP53 mutations than considering systemic DLBCL as a whole. Lauw et al. have suggested a higher prevalence of TP53 mutations and lower percentage of mutations in genes involved in BCR/TLR/ NF-κB signaling (e.g. MYD88, CARD11, CD79B, PIM1) in pediatric than adult CNS-DLBCL (135).
MYC, BCL2 and BCL6 rearrangements
In agreement with the ABC phenotype, chromosomal translocations affecting MYC and BCL2 gene are uncommon (25,35,36,39,40,74,91,136-139). However, CNS-DLBCL often overexpress MYC and BCL2 proteins (29,31,32,36,38-40,91,138), and gains or amplifications of the genomic loci are detected in 10–30% (25,36,39) and 20–40% (25,39,40,58,75), respectively (74).
Differently from MYC and BCL2, BCL6 is recurrently translocated in CNS-DLBCL with frequencies ranging from 17% to 44% (25,34,39,40,58,74,91,99,100,137-139), perhaps more commonly than what seen in the systemic ABC DLBCL C5/MCD/MYD88 cluster (47-50).
The ETS transcription factor ETV6 is frequently inactivated
The ETS transcription factor ETV6 appears inactivated by multiple mechanisms in CNS-DLBCL (57,58,93) as well as in systemic C5/MCD/MYD88 cluster (47-50). The gene is targeted by mutations in 16% (6/37) (57) and deletions in exons 2 or exons 2–5 in 13% (3/24) (58). Finally, the gene is translocated in the ETV6-IGH fusions in 18% (13/72) of the cases, with breakpoints leading to a truncated ETV6, lacking exons 1–2 (93). Albeit ETV6 is known to be involved in B-cell development, the contribution to CNS-DLBCL pathogenesis of its inactivation is still to be defined.
Different potential mechanism of immune evasion
Immune escape by loss of expression of MHC class I is a fundamental mechanism in DLBCL (140). Genetic events reported in systemic DLBCL and indeed enriched in the C5/MCD/MYD88 cluster (47-50,141) as mechanism to evade the immune system are common in CNS-DLBCL: B2M, CD58, CIITA are often targeted by somatic mutations, deletions or genomic rearrangements (15,56,58,59,78,81,85-87,94).
Integrating CNV and WES Braggio et al. reported inactivation of HLA genes by somatic mutations in 50% and B2M inactivation in approximately 10% of the cases (56). In a series of 19 CNS-DLBCL cases, Schwindt reported DNA losses or uniparental disomy in 74% of the cases at 6p21.32, the region spanning the MHC class I and II encoding genes (78). Fukumura et al. reported mutations in CD58 in 17%, B2M in 10%, CIITA (17%), HLA-C (15%), and focal deletions of HLA locus in one case (15).
The relevance of PDL1/PDL2 in CNS-DLBCL is undefined. Chapuy et al. reported chromosomal translocations the PDL1 locus in 3/24 cases (13%) (58). Genomic amplifications of PDL1/PDL2 are uncommon (25,69,71,78,142), and even their effect is uncertain since Chapuy et al. but not Villa et al. have reported concomitant protein over-expression in cases with genomic gain of the locus (25,58).
Finally, Waldera-Lupa et al. have studied CSF proteome in 17 CNS-DLBCL compared to 10 non-tumor samples (143). HLA class II HLA-DRB1 was among the proteins increased in CNS-DLBCL patients, suggesting that shedding of the detected protein could represent a mechanism of immune evasion (143).
miRNAs and CNS-DLBCL
Despite the rarity of the disease, quite a few studies have been published regarding the potential role of miRNAs (144-153). Known oncomirs such the members of the miR-17–92 cluster, in particular miR-17-5p, miR-155, miR-21 miR-196b, have been reported upregulated in CNS-DLBCL, similarly to other lymphoma subtypes (144-146,149-151). Unfortunately, due to the still uncharacterized function of most miRNAs, the results of the studies, reviewed elsewhere (154), are largely preliminary and descriptive.
Takashima et al. have presented a prognosis prediction model based on four miRNAs (miR-101, miR-548b, miR-554, and miR-1202), identified profiling 40 CNS-DLBCL focusing on miRNAs that regulate immunity (147).
Interestingly, miR-21, miR-19b, and miR-92a are more expressed in the CSF of patients with CNS-DLBCL than non-malignant brain lesions and might have a biomarker role (146,149-151), as also proposed for the U2 small nuclear RNA fragments (RNU2-1f), also detectable at high levels in the CSF of CNS-DLBCL (155).
Genetic lesions and clinical outcome
There are no clear associations between the presence of any genetic lesion and clinical outcome. This is due to the sample size of the series, the heterogeneity of therapies, the frequency of the lesions, and the different methodologies to define the presence of absence of the genetic event.
Across a series of heterogeneous 54 CNS-DLBCL, Zheng et al. reported a worse outcome for the four patients with mutated CD79B versus the 12 wild type patients (70). An inferior PFS for mutated CD79B cases has been seen also in a series of 57 cases with a mutation frequency of 59% (30) but not in another series of 71 patients (67).
No prognostic impact has been reported for the presence of MYD88 mutations in different series (59,67,70,156).
Deletions at 6q have been associated with inferior outcome in multiple series, also including HD-MTX treated patients (59,89,91,92).
Loss at 6q22, alongside with the presence of BCL6 translocation, was associated with inferior outcome in a series of 75 cases of CNS-DLBCL (38% treated with HD-MTX) (91).
Other genetic lesions potentially affecting the outcome of CNS-DLBCL patients include CDKN2A homozygous losses (59), BCL6 translocations (91), HLA-C mutations (15).
Munch-Petersen et al. studied TP53 in 86 CNS-DLBCL patients. Although the 32 mutated cases (34%) did not show differences in clinical outcome, a worse PFS and OS was seen for the nine patients bearing a TP53 mutation supposed to directly affect the direct protein/DNA contact versus all the other patients treated with combination chemotherapy with or without rituximab (26).
DNA mutations can be used to track CNS-DLBCL cells
The analysis of circulating tumor DNA (ctDNA) represents an important tool in the diagnostic and follow-up settings (130,157-161). As for systemic disease, tumor mutations can be detected in serum- or plasma-derived ctDNA of CNS-DLBCL (51,62,64,161,162). Moreover, the analyzed ctDNA in CSF might be more effective in detecting the presence of tumor cells than flow cytometry or cytology (63,163-166), and could be used to follow the patient during and after treatment (164). In the context of a prospective study, Ferreri et al. have detected the MYD88 L265P mutation in the CSF of 26/36 (72%) newly diagnosed and relapsed CNS-DLBCL patients, respectively (130). The combination of CSF IL-10 levels and MYD88 L265P appeared to identify disease in the CSF of 21/24 (88%) patients with relapsing CNS-DLBCL (130). At the 2021 Meeting of the American Society of Hematology, Mutter et al. reported the analysis of 85 tumor biopsies, 131 plasma samples, and 62 CSF specimens from 92 CNS-DLBCL patients using ultrasensitive sequencing technologies such CAPP-Seq (cancer personalized profiling by deep sequencing) and PhasED-Seq (phased variant enrichment and detection sequencing) (161). Genetic lesions were detected in 78% of plasma samples and in 100% of CSF specimens, and the levels of ctDNA, at baseline and during treatment appeared to predict PFS and OS (161). Moreover, the data suggest that the use of plasma or CSF ctDNA could be implemented in the diagnostic workflow to possibly avoid surgical biopsies for a subset of patients (161).
Genetic and biologic features suggest targeted therapies
The enrichment of specific genetic and biologic features in CNS-DLBCL (Table 1) provide the rationale for potential targeted therapies (127,167-174), which have been explored in a series of phase I and II studies (Table 2).
Table 1
| Cell of origin | ABC-DLBCL |
|---|---|
| Genetic subtype | C5/MCD |
| Immunoglobulin genes rearrangement | Non-switched self-reactive somatically mutated immunoglobulins |
| Recurrent mutations | CD79B, MYD88, ETV6, PIM1, SLIT2, TERT (BCR/TLR/NF-κB), HLA-A, B2M, CD58 (immune escape), ETV6, TBL1XR1, PRDM1 (B cell differentiation), KMT2D (chromatin remodeling) |
| Recurrent genomic gains | +18q (BCL2, MALT1), +3q (NFKBIZ), +18p, +3p, +19q13.42, +19q |
| Recurrent genomic losses | -6p21.32 (MHC), -6q21 (PRDM1), -9p21.3 (CDKN2A) |
| Recurrent fusions and translocations | 3q27 (BCL6) |
CNS-DLBCL, central nervous system diffuse large B-cell lymphoma; ABC, activated B-cell like.
Table 2
| Clinical phase | Compound | Mechanism of action | Population (cases) | ORR | CRR | NCT Number | Status | Reference, when available |
|---|---|---|---|---|---|---|---|---|
| I | Ibrutinib* | BTK inhibitor, chemotherapy, anti-CD20 | R/R (n=9) | 89%* | 67%* | NCT02315326 | Active, not recruiting | (175) |
| I | Lenalidomide | IMID | R/R (n=8) | 88% | 12% | NCT01542918 | Completed | (123) |
| I | Ibrutinib in the DA-TEDDI-R regimen^ | BTK inhibitor, chemotherapy, anti-CD20 | R/R (n=13), 1st line (n=5) | 83% | 86% | NCT02203526 | Recruiting | (176) |
| I | Ibrutinib, rituximab, lenalidomide | BTK inhibitor, IMID, anti-CD20 | R/R PCNSL and SCNSL (n=14) | 78% | 28% | NCT03703167 | Recruiting | (177) |
| I | Ibrutinib | BTK inhibitor | R/R (n=13) | 77% | 38% | – | – | (94) |
| I/II | Copanlisib, ibrutinib | BTK inhibitor, PI3Kα/δ inhibitor | R/R (n=6) | 67% | 17% | NCT03581942 | Recruiting | (164) |
| II | Ibrutinib | BTK inhibitor | R/R (n=52) | 52% | 20% | NCT02542514 | Active, not recruiting | (178) |
| II | Temsirolimus | mTOR inhibitor | R/R (n=37) | 54% | 22% | NCT00942747 | Unknown | (179) |
| I | Pomalidomide, dexamethasone | IMID, steroid | R/R PCNSL or 1st line/RR IOL (n=25) | 48% | 32% | NCT01722305 | Completed | (180) |
| II | Lenalidomide, rituximab | IMID, anti-CD20 | R/R (n=50) | 36%$ | 29%$ | NCT01956695 | Completed | (181) |
| II | Buparlisib | Pan-Pi3K inhibitor | R/R PCNSL and SCNSL (n=4) | 25% | 0% | NCT02301364 | Completed | (182) |
| I | Bimiralisib | mTOR/PI3K inhibitor | R/R | n.a. | n.a. | NCT02669511 | Completed | – |
| I | Rituximab, lenalidomide, and nivolumab | IMID, anti-PD1, anti-CD20 | R/R | n.a. | n.a. | NCT03558750 | Terminated (low accrual) | – |
| I | Acalabrutinib, rituximab, durvalumab | BTK inhibitor, anti-PDL1, anti-CD20 | R/R | n.a. | n.a. | NCT04688151 | Not yet recruiting | – |
| I | Nivolumab, pomalidomide | Anti-PD1, IMID | R/R | – | – | NCT03798314 | Active, not recruiting | – |
| I | CA-4948 | IRAK4 inhibitor | R/R | n.a. | n.a. | NCT03328078 | Recruiting | (183) |
| I | PRT811 | PRMT5 inhibitor | R/R | n.a. | n.a. | NCT04089449 | Recruiting | – |
| I | Venetoclax, obinutuzumab | BCL2 inhibitor, anti-CD20 | R/R | n.a. | n.a. | NCT04073147 | Recruiting | – |
| I | Pevonedistat, irinotecan, temozolomide | NEDD8 inhibitor, chemotherapy | R/R | n.a. | n.a. | NCT03323034 | Recruiting | (184) |
| II | Pembrolizumab | Anti-PD1 | R/R | n.a. | n.a. | NCT02779101 | Unknown | – |
| II | Sintilimab | Anti-PD1 | R/R | n.a. | n.a. | NCT04052659 | Recruiting | – |
| II | Acalabrutinib | BTK inhibitor | R/R | n.a. | n.a. | NCT04548648 | Not yet recruiting | – |
| II | Camrelizumab | PD1 | R/R | n.a. | n.a. | NCT04070040 | Recruiting | – |
| II | IBER (ibrutinib, rituximab, ifosfamide, etoposide) followed by ibrutinib as maintenance | BTK inhibitor, anti-CD20, chemotherapy | R/R | n.a. | n.a. | NCT04066920 | Not yet recruiting | – |
| II | F520 | anti-PD1 | R/R | n.a. | n.a. | NCT04457869 | Not yet recruiting | – |
| II | Ibrutinib vs. lenalidomide in combination with MRE (methotrexate, rituximab, etoposide) | BTK inhibitor, IMID, anti-CD20, chemotherapy | R/R | n.a. | n.a. | NCT04129710 | Recruiting | – |
| II | Abemaciclib | CDK4/6 inhibitor | R/R | n.a. | n.a. | NCT03220646 | Recruiting | – |
| II | Ibrutinib, nivolumab | BTK inhibitor, anti-PD1 | R/R | n.a. | n.a. | NCT03770416 | Recruiting | (185) |
| II | ICP-022 | BTK inhibitor | R/R | n.a. | n.a. | NCT04438044 | Recruiting | – |
| II | Pembrolizumab | Anti-PD1 | R/R | n.a. | n.a. | NCT03255018 | Active, not recruiting | – |
| II | Nivolumab | Anti-PD1 | R/R | n.a. | n.a. | NCT02857426 | Active, not recruiting | – |
| II | Bimiralisib | mTOR/PI3K inhibitor | R/R | n.a. | n.a. | NCT03120000 | Withdrawn | – |
| I/II | Pembrolizumab, ibrutinib and rituximab | BTK inhibitor, anti-PD1, anti-CD20 | R/R | n.a. | n.a. | NCT04421560 | Recruiting | – |
| I | Acalabrutinib and durvalumab | BTK inhibitor, anti-PDL1 | R/R, 1st line | n.a. | n.a. | NCT04462328 | Not yet recruiting | – |
| I | TEDDI-R | BTK inhibitor, chemotherapy, anti-CD20 | 1st line (n=13) | 92% | 67% | NCT02203526 | Recruiting | (186) |
| I | Nivolumab | Anti-PD1 | 1st line | n.a. | n.a. | NCT04022980 | Recruiting | – |
| I/II | R2-MTX with lenalidomide maintenance | IMID, chemotherapy, anti-CD20 | 1st line | n.a. | n.a. | NCT04120350 | Recruiting | – |
| I/II | Lenalidomide and ibrutinib in association with rituximab-methotrexate procarbazine vincristin (R-MPV) | IMID, BTK inhibitor, chemotherapy, anti-CD20 | 1st line | n.a. | n.a. | NCT04446962 | Recruiting | – |
| II | Ibrutinib, methotrexate, temozolomide | BTK inhibitor, chemotherapy | 1st line | n.a. | n.a. | NCT04514393 | Not yet recruiting | – |
| II | Lenalidomide, rituximab, methotrexate | IMID, anti-CD20, chemotherapy | 1st line | n.a. | n.a. | NCT04481815 | Recruiting | – |
| II | Nivolumab, maintenance | Anti-PD1 | 1st line | n.a. | n.a. | NCT04401774 | Recruiting | – |
| II | Rituximab, lenalidomide combined with methotrexate and temozolomide | IMID, anti-CD20, chemotherapy | 1st line | n.a. | n.a. | NCT04737889 | Recruiting | – |
| II | Ibrutinib as maintenance | BTK inhibitor | 1st line | n.a. | n.a. | NCT02623010 | Recruiting | – |
| II | Lenalidomide, rituximab maintenance | IMID, anti-CD20 | 1st line | n.a. | n.a. | NCT04627753 | Recruiting | – |
| II | Lenalidomide and nivolumab to the usual treatment | IMID, anti-PD1 | 1st line | n.a. | n.a. | NCT04609046 | Recruiting | – |
| II | Chidamide, rituximab, methotrexate | HDAC inhibitor, anti-CD20, chemotherapy | 1st line | n.a. | n.a. | NCT04516655 | Not yet recruiting | – |
| II | Lenalidomide vs. procarbazine as maintenance after HDX-MTX-based regimen | IMID, chemotherapy | 1st line | n.a. | n.a. | NCT03495960 | Recruiting | – |
*, in combination with rituximab and high-dose methotrexate and as maintenance; ^, based on ibrutinib alone given for 14-day prior to cycle 1 of DA-TEDDI-R (temozolomide, etoposide, liposomal doxorubicin, dexamethasone, ibrutinib, and rituximab) with intraventricular cytarabine; $, on the 23 patients that received 8 cycles. CNS-DLBCL, central nervous system diffuse large B-cell lymphoma; ORR, overall response rate; R/R, refractory/relapsed; CRR, complete remission rate; IMID, immunomodulatory drug; n.a., not available.
CNS-DLBCL are highly enriched of MYD88 L265P and CD79B mutations that provide the strong rationale for testing BTK inhibitors (94,164,176,178,187). Indeed, in systemic ABC DLBCL, these genetic features have been associated with higher sensitivity to this therapeutic approach (188). Since CNS-DLBCL are highly enriched of these features, it made sense to test the use of the BTK inhibitor ibrutinib (94,164,176,178,187). Preclinical and clinical data show that ibrutinib passes the blood brain barrier (176,189,190), and the drug has been tested both as single agent (94) and in combination (164,176). Interestingly, the relationship between genetic status and clinical responses is intriguing with responses observed also in patients lacking MYD88/CD79B mutations, or in patients with tumors with genetic lesions, such as CARD11 mutations, expected to cause ibrutinib resistance (164,176,178). For example, in the study reported by Grommes et al., non-responders were enriched of lesions supposed to give resistance to ibrutinib (CARD11 mutations, TNFAIP3 inactivation), but, differently from systemic DLBCL (99), there were no complete remissions (CR) in patients with tumors bearing both MYD88 and CD79B mutations (three partial response and one stable disease) (94). Two of three CR cases had MYD88 L265P but lacked CD79B mutations and one was wild type for both MYD88 and CD79B (94).
In vitro evidence obtained in cell lines derived from the systemic ABC DLBCL C5/MCD/MYD88 cluster showed a strong synergism for the combination of ibrutinib with DNA damaging agents (doxorubicin, etoposide, cytarabine, and mitomycin C) and antagonism when the BTK inhibitor was combined with multiple anti-folates, apparently not supporting a combination with methotrexate-based regimens (176). However, clinically, a similar overall response rate (ORR) has been achieved with ibrutinib combined with both DNA damaging agents (176) and methotrexate-based regimens (164).
It must be mentioned that in the phase I combining ibrutinib with DNA damaging agents in 18 CNS-DLBCL patients, major infections were common: five cases of pulmonary aspergillosis, one case of pulmonary Pneumocystis jiroveci, and three pneumonitis of undetermined etiology, two cases of CNS aspergillosis, and one case of enterocolitis (176). Eight of the 18 patients died, three from disease progression, two due to Aspergillus infection during the ibrutinib window, and one due to neutropenic sepsis during chemotherapy (176). Preliminary data from the ongoing follow-up study suggest that the introduction of fungal prophylaxis seems to avoid aspergillosis (186).
Clinical studies are now exploring BTK inhibitors also in combination with additional targeted agents such immunomodulatory drugs (IMiDs), anti-CD20 monoclonal antibodies against CD20 or checkpoint modulators. Among the first 15 patients (11 with CNS-DLBCL, four with secondary CNS lymphoma) enrolled in a phase I study combining ibrutinib, with the IMID lenalidomide and the anti-CD20 monoclonal antibody rituximab, no Aspergillosis infection was observed, with thrombocytopenia, lymphopenia, and rash as the most adverse events (177).
In addition to an active BTK signaling, CNS-DLBCL cells also depend on the PI3K/AKT/mTOR and RAS/MAPK signaling (94,191-193). Active mTOR signaling has been reported in over 70% of 24 CNS-DLBCL cases, using phospho-4E-BP1 (Thr 37/46) and phospho-S6 (Ser235/236) as downstream markers of activity (192). The transcriptome of CNS-DLBCL cases with CD79B mutations is enriched of gene expression signatures associated with PI3K and mTOR signaling, while NFKB signatures are under-represented (94). The need of targeting of multiple proteins is suggested also by the preclinical observation that the killing of CD79B-mutant CNS-DLBCL patient-derived xenografts is not achieved using ibrutinib or isoform specific PI3K inhibitors as single agents, but only with pan PI3K inhibitors, dual PI3Kα/δ inhibition, or combining low concentrations of the BTK inhibitor with dual PI3Kα/δ or mTOR inhibitors (94). Phase I–II studies performed in the relapsed/refractory (R/R) setting have so far reported only modest single agent activity with the mTOR inhibitor temsirolimus (179) or the pan-PI3K inhibitor buparlisib (182). No data have yet been reported with the dual PI3K/mTOR inhibitor bimiralisib. Albeit based on the very first six patients treated in the escalation phase (164), the combination of ibrutinib and the PI3Kα/δ inhibitor copanlisib seems safe but not associated with much better results, at least in terms of response rate, than what seen with single BTK inhibition. It must also be mentioned that the observed phospho-S6 might be downstream to an additional kinase, PASK, and not mTOR, suggesting different therapeutic targets (193).
The typical MCD phenotype of CNS-DLBCL cells, the role of IMIDS in targeting the IRF4/SPIB axis in systemic MCD DLBCL models as well as the reported synergisms with IMDS and ibrutinib (194) represent the rationale to the clinical studies with lenalidomide or pomalidomide as single agents and in combination (123,177,180,181).
There are a whole lot of additional targets that have not yet been clinically explored in the CNS-DLBCL setting.
Jiménez et al. reported preclinical synergism combining ibrutinib with selinexor, a selective inhibitor of nuclear export (SINE) compound that specifically blocks Exportin 1 (XPO1) (195). The combination of the two small molecules has a direct anti-tumor effect but it also able to modulate the tumor microenvironment pushing toward a pro-inflammatory M1-like macrophages and reducing PD1 and SIRPα expression on M2-like macrophages (195).
Using orthotopic patient derived xenografts, Tateishi et al. have shown the therapeutic potential of the direct NF-κB inhibition using BAY11-7082 and of the natural compound juglone, an inhibitor of peptidyl-prolyl isomerase PIN1, as anti-lymphoma agents and to sensitize cells to chemotherapy (16). Since NF-κB activation has been associated to low sensitivity to chemotherapy, including high-dose (HD) methotrexate (MTX) (16), inhibition of the pathway, as also achieved with BTK inhibitors, might improve the patient’s outcome.
The frequent inactivation of CDKN2A and the high genomic instability suggest the possible benefit of using aurora kinase inhibitors, CDK4/6 inhibitors (196) or PRMT5 inhibitors (197).
As previously mentioned, CNS-DLBCL cells from different patients still recognize the same antigens, and in particular SAMD14 and neurabin-I (11). Bewarder et al. applied the B-cell receptor Antigen for Reverse targeting (BAR) approach to develop a molecule that resembles an IgG1 antibody and contains the BCR-binding epitope of the common CNS-DLBCL antigens SAMD14 and neurabin-I instead of variable regions to target the B-cell receptors with specificity for neurabin-I (198). The IgG1-format neurabin-I BAR-body shows in vitro ability to induce antibody-dependent cell-mediated cytotoxicity and, based on in vivo data obtained with a similar molecule, it might represent a future therapeutic approach (199), although its efficacy and safety still require extensive validations (200).
Due to the expression of BAFF-R on CNS-DLBCL cells and on the role of BAFF signaling in sustaining their growth, antibodies against the soluble BAFF of against BAFF-R have been proposed as potential therapeutic tools for CNS-DLBCL patients (109).
Promoter methylation affecting specific genes can predict the response to specific drugs. Hypermethylation of O6-methylguanine- DNA methyltransferase (MGMT) promoter region has been reported in 30-60% of CNS-DLBCL cases (70,77,84,201,202) and suggests a higher sensitivity to alkylating agents as reported for temozolomide or carmustine (BCNU) in glioma patients (203,204). In five relapsed CNS-DLBCL patients treated with temozolomide, the four responders had methylated MGMT promoter, while the only non-responders had no methylation (202). In a series of 15 CNS-DLBCL cases treated with temozolomide and analyzed for MGMT protein expression, all patients with low expression achieved CR or PR and only one with high MGMT expression responded to treatment (205).
Inversely to MGMT, methylation in the promoter of the gene coding for RFC (reduced folate carrier), involved in MTX cellular uptake, seems to be associated with decreased response to HD-MTX (206). In a series of 37 CNS-DLBCL patients, RFC promoter methylation was detected in 24% of the cases (206). None of patients with promoter methylation were relapse-free at three years and all the patients alive at 3 years had unmethylated RFC (206).
Somatic mutations in the CD19 gene have not yet been reported in CNS-DLBCL patients but the increase in use of anti-CD19 chimeric antigen-receptor (CAR) T-cells in these patients population (Table 3), sustained by initial promising reports (207-209), will require its inclusion especially in targeted DNA sequencing, especially in follow-up analyses.
Table 3
| Clinical phase | Compound | Mechanism of action | Population | ORR | CRR | NCT number | Status |
|---|---|---|---|---|---|---|---|
| I | CD19CAT-41BBζ CAR T-cells | Anti-CD19 CAR T-cells | R/R | n.a. | n.a. | NCT04443829 | Recruiting |
| I | Tisagenlecleucel | Anti-CD19 CAR T-cells | R/R, 1st line | n.a. | n.a. | NCT04134117 | Recruiting |
| I | Axicabtagene ciloleucel | Anti-CD19 CAR T-cells | R/R | n.a. | n.a. | NCT04608487 | Recruiting |
| I | 19(T2)28z1xx CAR T cells | Anti-CD19 CAR T-cells | R/R | n.a. | n.a. | NCT04464200 | Recruiting |
| I | CAR T-cells | Anti-CD19 CAR T-cells | R/R | n.a. | n.a. | NCT04532203 | Recruiting |
| II | Lisocabtagene maraleucel | Anti-CD19 CAR T-cells | R/R | n.a. | n.a. | NCT03484702 | Recruiting |
ORR, overall response rate; R/R, refractory/relapsed; CRR, complete remission rate; CAR, chimeric antigen-receptor.
It is important to highlight that mutation/methylation screening approach does not identify all the potential therapeutic targets by ignoring surrogate marker. These markers are always expressed by the tumor cells (but not exclusively) and with CD20 as one example are already used as target molecule in therapies. another surrogate marker in CNS-DLBCL is CXCR4, which is always expressed by the tumor cells (210), and never reported mutated in the setting of CNS-DLBCL (211). The ligand CXCL12 is expressed by endothelial cells, microglia cells (only in the infiltration zone) and by the tumor cells itself (210). Perhaps it provides a crucial signal leading to a permissive tumor microenvironment and immune evasion, as described for other cancer types (212), and its targeting might be helpful. Future therapy concepts will indeed focus on personalized targeting. Brain biopsy remains the gold standard for the diagnostic of CNS-DLBCL and it will become the foundation for clinical decisions, although it is important to remind that no single CNS-DLBCL biopsy contains all the hereby described genetic aberrations (Figure 2).
In conclusion, our knowledge on the genetic and epigenetic alteration sustaining CNS-DLBCL cells has increased, strengthening the diagnostic criteria and making this disorder a more homogenous disease. Its close link with the systemic MCD/C5 genetic clusters and specific biologic features now provides new opportunities. Novel agents with the capacity to cross the BBB and targeting CNS-DLBCL cells or their tumor microenvironment can be tested and included in future therapeutic schemes. Innovative follow-up approaches can be explored with the use of liquid biopsies and the detection of secreted factors in serum/plasma and/or CSF.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Andrés J. M. Ferreri and Maurilio Ponzoni) for the series “Central Nervous System Lymphomas” published in Annals of Lymphoma. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://aol.amegroups.com/article/view/10.21037/aol-21-38/coif). The series “Central Nervous System Lymphomas” was commissioned by the editorial office without any funding or sponsorship. FB reports institutional research funds from ADC Therapeutics, Bayer AG, Helsinn, Curis, Menarini Ricerche, NEOMED Therapeutics 1, Nordic Nanovector ASA, and Polyphor; travel grant from Astra Zeneca, Consulting fees from Menarini Ricerche. MMR reports personal payment from Deutsche Krebshilfe, Wilhelm Sander-Stiftung, Marga und Walter Boll-Stiftung, Köln Fortune, and institutional payment from Deutsche Forschungsgemeinschaft. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Batchelor TT. Primary central nervous system lymphoma: A curable disease. Hematol Oncol 2019;37:15-8. [Crossref] [PubMed]
- Kluin PM, Deckert M, Ferry JA. Primary diffuse large B-cell lymphoma of the CNS. In: Swerdlow SH, Campo E, Harris NL, et al. (editors). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues - Revised 4th Edition. Lyon, France: IARC Press, 2017:300-2.
- Gandhi MK, Hoang T, Law SC, et al. EBV-associated primary CNS lymphoma occurring after immunosuppression is a distinct immunobiological entity. Blood 2021;137:1468-77. [Crossref] [PubMed]
- Montesinos-Rongen M, Küppers R, Schlüter D, et al. Primary central nervous system lymphomas are derived from germinal-center B cells and show a preferential usage of the V4-34 gene segment. Am J Pathol 1999;155:2077-86. [Crossref] [PubMed]
- Thompsett AR, Ellison DW, Stevenson FK, et al. V(H) gene sequences from primary central nervous system lymphomas indicate derivation from highly mutated germinal center B cells with ongoing mutational activity. Blood 1999;94:1738-46. [Crossref] [PubMed]
- Sekita T, Tamaru JI, Kaito K, et al. Primary central nervous system lymphomas express Vh genes with intermediate to high somatic mutations. Leuk Lymphoma 2001;41:377-85. [Crossref] [PubMed]
- Montesinos-Rongen M, Purschke F, Küppers R, et al. Immunoglobulin repertoire of primary lymphomas of the central nervous system. J Neuropathol Exp Neurol 2014;73:1116-25. [Crossref] [PubMed]
- Pels H, Montesinos-Rongen M, Schaller C, et al. VH gene analysis of primary CNS lymphomas. J Neurol Sci 2005;228:143-7. [Crossref] [PubMed]
- Montesinos-Rongen M, Purschke FG, Brunn A, et al. Primary Central Nervous System (CNS) Lymphoma B Cell Receptors Recognize CNS Proteins. J Immunol 2015;195:1312-9. [Crossref] [PubMed]
- Vater I, Montesinos-Rongen M, Schlesner M, et al. The mutational pattern of primary lymphoma of the central nervous system determined by whole-exome sequencing. Leukemia 2015;29:677-85. [Crossref] [PubMed]
- Thurner L, Preuss KD, Bewarder M, et al. Hyper-N-glycosylated SAMD14 and neurabin-I as driver autoantigens of primary central nervous system lymphoma. Blood 2018;132:2744-53. [Crossref] [PubMed]
- Malumbres R, Davis J, Ruiz P, et al. Somatically mutated immunoglobulin IGHV@ genes without intraclonal heterogeneity indicate a postgerminal centre origin of primary intraocular diffuse large B-cell lymphomas. Br J Haematol 2007;138:749-55. [Crossref] [PubMed]
- Montesinos-Rongen M, Schmitz R, Courts C, et al. Absence of immunoglobulin class switch in primary lymphomas of the central nervous system. Am J Pathol 2005;166:1773-9. [Crossref] [PubMed]
- Montesinos-Rongen M, Van Roost D, Schaller C, et al. Primary diffuse large B-cell lymphomas of the central nervous system are targeted by aberrant somatic hypermutation. Blood 2004;103:1869-75. [Crossref] [PubMed]
- Fukumura K, Kawazu M, Kojima S, et al. Genomic characterization of primary central nervous system lymphoma. Acta Neuropathol 2016;131:865-75. [Crossref] [PubMed]
- Tateishi K, Miyake Y, Kawazu M, et al. A Hyperactive RelA/p65-Hexokinase 2 Signaling Axis Drives Primary Central Nervous System Lymphoma. Cancer Res 2020;80:5330-43. [Crossref] [PubMed]
- Cascione L, Aresu L, Baudis M, et al. DNA Copy Number Changes in Diffuse Large B Cell Lymphomas. Front Oncol 2020;10:584095. [Crossref] [PubMed]
- Swerdlow SH, Campo E, Harris NL, et al. (editors). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues - Revised 4th Edition. World Health Organization Classification of Tumours. Lyon, France: IARC Press, 2017.
- Pasqualucci L. Molecular pathogenesis of germinal center-derived B cell lymphomas. Immunol Rev 2019;288:240-61. [Crossref] [PubMed]
- Miao Y, Medeiros LJ, Li Y, et al. Genetic alterations and their clinical implications in DLBCL. Nat Rev Clin Oncol 2019;16:634-52. [Crossref] [PubMed]
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503-11. [Crossref] [PubMed]
- Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002;346:1937-47. [Crossref] [PubMed]
- Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 2008;359:2313-23. [Crossref] [PubMed]
- Lenz G, Wright GW, Emre NC, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A 2008;105:13520-5. [Crossref] [PubMed]
- Villa D, Tan KL, Steidl C, et al. Molecular features of a large cohort of primary central nervous system lymphoma using tissue microarray. Blood Adv 2019;3:3953-61. [Crossref] [PubMed]
- Munch-Petersen HD, Asmar F, Dimopoulos K, et al. TP53 hotspot mutations are predictive of survival in primary central nervous system lymphoma patients treated with combination chemotherapy. Acta Neuropathol Commun 2016;4:40. [Crossref] [PubMed]
- Hatzl S, Posch F, Deutsch A, et al. Immunohistochemistry for c-myc and bcl-2 overexpression improves risk stratification in primary central nervous system lymphoma. Hematol Oncol 2020;38:277-83. [Crossref] [PubMed]
- Camilleri-Broët S, Crinière E, Broët P, et al. A uniform activated B-cell-like immunophenotype might explain the poor prognosis of primary central nervous system lymphomas: analysis of 83 cases. Blood 2006;107:190-6. [Crossref] [PubMed]
- Shi QY, Feng X, Bao W, et al. MYC/BCL2 Co-Expression Is a Stronger Prognostic Factor Compared With the Cell-of-Origin Classification in Primary CNS DLBCL. J Neuropathol Exp Neurol 2017;76:942-8. [Crossref] [PubMed]
- Zhou Y, Liu W, Xu Z, et al. Analysis of Genomic Alteration in Primary Central Nervous System Lymphoma and the Expression of Some Related Genes. Neoplasia 2018;20:1059-69. [Crossref] [PubMed]
- Kim S, Nam SJ, Kwon D, et al. MYC and BCL2 overexpression is associated with a higher class of Memorial Sloan-Kettering Cancer Center prognostic model and poor clinical outcome in primary diffuse large B-cell lymphoma of the central nervous system. BMC Cancer 2016;16:363. [Crossref] [PubMed]
- Chen Y, Chen H, Chen L, et al. Immunohistochemical overexpression of BCL-2 protein predicts an inferior survival in patients with primary central nervous system diffuse large B-cell lymphoma. Medicine (Baltimore) 2019;98:e17827. [Crossref] [PubMed]
- Bödör C, Alpár D, Marosvári D, et al. Molecular Subtypes and Genomic Profile of Primary Central Nervous System Lymphoma. J Neuropathol Exp Neurol 2020;79:176-83. [Crossref] [PubMed]
- Preusser M, Woehrer A, Koperek O, et al. Primary central nervous system lymphoma: a clinicopathological study of 75 cases. Pathology 2010;42:547-52. [Crossref] [PubMed]
- Son SM, Ha SY, Yoo HY, et al. Prognostic impact of MYC protein expression in central nervous system diffuse large B-cell lymphoma: comparison with MYC rearrangement and MYC mRNA expression. Mod Pathol 2017;30:4-14. [Crossref] [PubMed]
- Gill KZ, Iwamoto F, Allen A, et al. MYC protein expression in primary diffuse large B-cell lymphoma of the central nervous system. PLoS One 2014;9:e114398. [Crossref] [PubMed]
- Lim DH, Kim WS, Kim SJ, et al. Microarray Gene-expression Profiling Analysis Comparing PCNSL and Non-CNS Diffuse Large B-Cell Lymphoma. Anticancer Res 2015;35:3333-40. [PubMed]
- Makino K, Nakamura H, Shinojima N, et al. BCL2 expression is associated with a poor prognosis independent of cellular origin in primary central nervous system diffuse large B-cell lymphoma. J Neurooncol 2018;140:115-21. [Crossref] [PubMed]
- Tapia G, Baptista MJ, Muñoz-Marmol AM, et al. MYC protein expression is associated with poor prognosis in primary diffuse large B-cell lymphoma of the central nervous system. APMIS 2015;123:596-603. [Crossref] [PubMed]
- Yin W, Xia X, Wu M, et al. The impact of BCL-2/MYC protein expression and gene abnormality on primary central nervous system diffuse large B-cell lymphoma. Int J Clin Exp Pathol 2019;12:2215-23. [PubMed]
- Inoue A, Ohnishi T, Kohno S, et al. Prognostic significance of immunohistochemical subtypes based on the stage of B-cell differentiation in primary CNS lymphoma. Int J Clin Exp Pathol 2019;12:1457-67. [Crossref] [PubMed]
- Raoux D, Duband S, Forest F, et al. Primary central nervous system lymphoma: immunohistochemical profile and prognostic significance. Neuropathology 2010;30:232-40. [Crossref] [PubMed]
- Kawaguchi A, Iwadate Y, Komohara Y, et al. Gene expression signature-based prognostic risk score in patients with primary central nervous system lymphoma. Clin Cancer Res 2012;18:5672-81. [Crossref] [PubMed]
- Montesinos-Rongen M, Brunn A, Bentink S, et al. Gene expression profiling suggests primary central nervous system lymphomas to be derived from a late germinal center B cell. Leukemia 2008;22:400-5. [Crossref] [PubMed]
- Rubenstein JL, Fridlyand J, Shen A, et al. Gene expression and angiotropism in primary CNS lymphoma. Blood 2006;107:3716-23. [Crossref] [PubMed]
- Hilal T, Maguire A, Kosiorek HE, et al. Clinical features and cell of origin subtyping using gene expression profiling in HIV-negative patients with primary central nervous system lymphoma. Leuk Lymphoma 2019;60:3581-3. [Crossref] [PubMed]
- Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018;24:679-90. [Crossref] [PubMed]
- Schmitz R, Wright GW, Huang DW, et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med 2018;378:1396-407. [Crossref] [PubMed]
- Wright GW, Huang DW, Phelan JD, et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications. Cancer Cell 2020;37:551-568.e14. [Crossref] [PubMed]
- Lacy SE, Barrans SL, Beer PA, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood 2020;135:1759-71. [Crossref] [PubMed]
- Fontanilles M, Marguet F, Bohers É, et al. Non-invasive detection of somatic mutations using next-generation sequencing in primary central nervous system lymphoma. Oncotarget 2017;8:48157-68. [Crossref] [PubMed]
- Arai A, Takase H, Yoshimori M, et al. Gene expression profiling of primary vitreoretinal lymphoma. Cancer Sci 2020;111:1417-21. [Crossref] [PubMed]
- Barakat M, Albitar M, Whitney R, et al. Diversity of genetic alterations of primary central nervous system lymphoma in Hispanic versus non-Hispanic patients. Cancer Treat Res Commun 2021;27:100310. [Crossref] [PubMed]
- Bonzheim I, Giese S, Deuter C, et al. High frequency of MYD88 mutations in vitreoretinal B-cell lymphoma: a valuable tool to improve diagnostic yield of vitreous aspirates. Blood 2015;126:76-9. [Crossref] [PubMed]
- Zorofchian S, El-Achi H, Yan Y, et al. Characterization of genomic alterations in primary central nervous system lymphomas. J Neurooncol 2018;140:509-17. [Crossref] [PubMed]
- Braggio E, Van Wier S, Ojha J, et al. Genome-Wide Analysis Uncovers Novel Recurrent Alterations in Primary Central Nervous System Lymphomas. Clin Cancer Res 2015;21:3986-94. [Crossref] [PubMed]
- Bruno A, Boisselier B, Labreche K, et al. Mutational analysis of primary central nervous system lymphoma. Oncotarget 2014;5:5065-75. [Crossref] [PubMed]
- Chapuy B, Roemer MG, Stewart C, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood 2016;127:869-81. [Crossref] [PubMed]
- Gonzalez-Aguilar A, Idbaih A, Boisselier B, et al. Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin Cancer Res 2012;18:5203-11. [Crossref] [PubMed]
- Hattori K, Sakata-Yanagimoto M, Kusakabe M, et al. Genetic evidence implies that primary and relapsed tumors arise from common precursor cells in primary central nervous system lymphoma. Cancer Sci 2019;110:401-7. [Crossref] [PubMed]
- Hattori K, Sakata-Yanagimoto M, Okoshi Y, et al. MYD88 (L265P) mutation is associated with an unfavourable outcome of primary central nervous system lymphoma. Br J Haematol 2017;177:492-4. [Crossref] [PubMed]
- Hattori K, Sakata-Yanagimoto M, Suehara Y, et al. Clinical significance of disease-specific MYD88 mutations in circulating DNA in primary central nervous system lymphoma. Cancer Sci 2018;109:225-30. [Crossref] [PubMed]
- Hiemcke-Jiwa LS, Leguit RJ, Snijders TJ, et al. MYD88 p.(L265P) detection on cell-free DNA in liquid biopsies of patients with primary central nervous system lymphoma. Br J Haematol 2019;185:974-7. [Crossref] [PubMed]
- Montesinos-Rongen M, Brunn A, Tuchscherer A, et al. Analysis of Driver Mutational Hot Spots in Blood-Derived Cell-Free DNA of Patients with Primary Central Nervous System Lymphoma Obtained before Intracerebral Biopsy. J Mol Diagn 2020;22:1300-7. [Crossref] [PubMed]
- Takashima Y, Sasaki Y, Hayano A, et al. Target amplicon exome-sequencing identifies promising diagnosis and prognostic markers involved in RTK-RAS and PI3K-AKT signaling as central oncopathways in primary central nervous system lymphoma. Oncotarget 2018;9:27471-86. [Crossref] [PubMed]
- Montesinos-Rongen M, Godlewska E, Brunn A, et al. Activating L265P mutations of the MYD88 gene are common in primary central nervous system lymphoma. Acta Neuropathol 2011;122:791-2. [Crossref] [PubMed]
- Nakamura T, Tateishi K, Niwa T, et al. Recurrent mutations of CD79B and MYD88 are the hallmark of primary central nervous system lymphomas. Neuropathol Appl Neurobiol 2016;42:279-90. [Crossref] [PubMed]
- Narasimhan S, Joshi M, Parameswaran S, et al. MYD88 L265P mutation in intraocular lymphoma: A potential diagnostic marker. Indian J Ophthalmol 2020;68:2160-5. [Crossref] [PubMed]
- Nayyar N, White MD, Gill CM, et al. MYD88 L265P mutation and CDKN2A loss are early mutational events in primary central nervous system diffuse large B-cell lymphomas. Blood Adv 2019;3:375-83. [Crossref] [PubMed]
- Zheng M, Perry AM, Bierman P, et al. Frequency of MYD88 and CD79B mutations, and MGMT methylation in primary central nervous system diffuse large B-cell lymphoma. Neuropathology 2017;37:509-16. [Crossref] [PubMed]
- Ou A, Sumrall A, Phuphanich S, et al. Primary CNS lymphoma commonly expresses immune response biomarkers. Neurooncol Adv 2020;2:vdaa018.
- Kaulen LD, Erson-Omay EZ, Henegariu O, et al. Exome sequencing identifies SLIT2 variants in primary CNS lymphoma. Br J Haematol 2021;193:375-9. [Crossref] [PubMed]
- Montesinos-Rongen M, Schäfer E, Siebert R, et al. Genes regulating the B cell receptor pathway are recurrently mutated in primary central nervous system lymphoma. Acta Neuropathol 2012;124:905-6. [Crossref] [PubMed]
- Montesinos-Rongen M, Zühlke-Jenisch R, Gesk S, et al. Interphase cytogenetic analysis of lymphoma-associated chromosomal breakpoints in primary diffuse large B-cell lymphomas of the central nervous system. J Neuropathol Exp Neurol 2002;61:926-33. [Crossref] [PubMed]
- Harada K, Nishizaki T, Kubota H, et al. Distinct primary central nervous system lymphoma defined by comparative genomic hybridization and laser scanning cytometry. Cancer Genet Cytogenet 2001;125:147-50. [Crossref] [PubMed]
- Nakamura M, Sakaki T, Hashimoto H, et al. Frequent alterations of the p14(ARF) and p16(INK4a) genes in primary central nervous system lymphomas. Cancer Res 2001;61:6335-9. [PubMed]
- Cobbers JM, Wolter M, Reifenberger J, et al. Frequent inactivation of CDKN2A and rare mutation of TP53 in PCNSL. Brain Pathol 1998;8:263-76. [Crossref] [PubMed]
- Schwindt H, Vater I, Kreuz M, et al. Chromosomal imbalances and partial uniparental disomies in primary central nervous system lymphoma. Leukemia 2009;23:1875-84. [Crossref] [PubMed]
- Li X, Huang Y, Bi C, et al. Primary central nervous system diffuse large B-cell lymphoma shows an activated B-cell-like phenotype with co-expression of C-MYC, BCL-2, and BCL-6. Pathol Res Pract 2017;213:659-65. [Crossref] [PubMed]
- Wang L, Sato-Otsubo A, Sugita S, et al. High-resolution genomic copy number profiling of primary intraocular lymphoma by single nucleotide polymorphism microarrays. Cancer Sci 2014;105:592-9. [Crossref] [PubMed]
- Weber T, Weber RG, Kaulich K, et al. Characteristic chromosomal imbalances in primary central nervous system lymphomas of the diffuse large B-cell type. Brain Pathol 2000;10:73-84. [Crossref] [PubMed]
- Hayashi Y, Iwato M, Arakawa Y, et al. Homozygous deletion of INK4a/ARF genes and overexpression of bcl-2 in relation with poor prognosis in immunocompetent patients with primary central nervous system lymphoma of the diffuse large B-cell type. J Neurooncol 2001;55:51-8. [Crossref] [PubMed]
- Braaten KM, Betensky RA, de Leval L, et al. BCL-6 expression predicts improved survival in patients with primary central nervous system lymphoma. Clin Cancer Res 2003;9:1063-9. [PubMed]
- Chu LC, Eberhart CG, Grossman SA, et al. Epigenetic silencing of multiple genes in primary CNS lymphoma. Int J Cancer 2006;119:2487-91. [Crossref] [PubMed]
- Booman M, Szuhai K, Rosenwald A, et al. Genomic alterations and gene expression in primary diffuse large B-cell lymphomas of immune-privileged sites: the importance of apoptosis and immunomodulatory pathways. J Pathol 2008;216:209-17. [Crossref] [PubMed]
- Sung CO, Kim SC, Karnan S, et al. Genomic profiling combined with gene expression profiling in primary central nervous system lymphoma. Blood 2011;117:1291-300. [Crossref] [PubMed]
- Braggio E, McPhail ER, Macon W, et al. Primary central nervous system lymphomas: a validation study of array-based comparative genomic hybridization in formalin-fixed paraffin-embedded tumor specimens. Clin Cancer Res 2011;17:4245-53. [Crossref] [PubMed]
- Courts C, Montesinos-Rongen M, Brunn A, et al. Recurrent inactivation of the PRDM1 gene in primary central nervous system lymphoma. J Neuropathol Exp Neurol 2008;67:720-7. [Crossref] [PubMed]
- Nakamura M, Kishi M, Sakaki T, et al. Novel tumor suppressor loci on 6q22-23 in primary central nervous system lymphomas. Cancer Res 2003;63:737-41. [PubMed]
- Boonstra R, Koning A, Mastik M, et al. Analysis of chromosomal copy number changes and oncoprotein expression in primary central nervous system lymphomas: frequent loss of chromosome arm 6q. Virchows Arch 2003;443:164-9. [Crossref] [PubMed]
- Cady FM, O'Neill BP, Law ME, et al. Del(6)(q22) and BCL6 rearrangements in primary CNS lymphoma are indicators of an aggressive clinical course. J Clin Oncol 2008;26:4814-9. [Crossref] [PubMed]
- McPhail ER, Law ME, Decker PA, et al. Influence of 6q22-23 on overall survival in primary central nervous system lymphoma. Analysis of North Central Cancer Treatment Group trials 86 72 52, 93 73 51 and 96 73 51. Br J Haematol 2011;154:146-50. [Crossref] [PubMed]
- Bruno A, Labreche K, Daniau M, et al. Identification of novel recurrent ETV6-IgH fusions in primary central nervous system lymphoma. Neuro Oncol 2018;20:1092-100. [Crossref] [PubMed]
- Grommes C, Pastore A, Palaskas N, et al. Ibrutinib Unmasks Critical Role of Bruton Tyrosine Kinase in Primary CNS Lymphoma. Cancer Discov 2017;7:1018-29. [Crossref] [PubMed]
- Shah N, Pang B, Yeoh KG, et al. Potential roles for the PIM1 kinase in human cancer - a molecular and therapeutic appraisal. Eur J Cancer 2008;44:2144-51. [Crossref] [PubMed]
- Brault L, Gasser C, Bracher F, et al. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica 2010;95:1004-15. [Crossref] [PubMed]
- Baron BW, Anastasi J, Hyjek EM, et al. PIM1 gene cooperates with human BCL6 gene to promote the development of lymphomas. Proc Natl Acad Sci U S A 2012;109:5735-9. [Crossref] [PubMed]
- Szydłowski M, Prochorec-Sobieszek M, Szumera-Ciećkiewicz A, et al. Expression of PIM kinases in Reed-Sternberg cells fosters immune privilege and tumor cell survival in Hodgkin lymphoma. Blood 2017;130:1418-29. [Crossref] [PubMed]
- Montesinos-Rongen M, Akasaka T, Zühlke-Jenisch R, et al. Molecular characterization of BCL6 breakpoints in primary diffuse large B-cell lymphomas of the central nervous system identifies GAPD as novel translocation partner. Brain Pathol 2003;13:534-8. [Crossref] [PubMed]
- Schwindt H, Akasaka T, Zühlke-Jenisch R, et al. Chromosomal translocations fusing the BCL6 gene to different partner loci are recurrent in primary central nervous system lymphoma and may be associated with aberrant somatic hypermutation or defective class switch recombination. J Neuropathol Exp Neurol 2006;65:776-82. [Crossref] [PubMed]
- Mondello P, Tarantelli C, Cascione L, et al. Inhibition of PIM Kinases Targets Synthetic Vulnerabilities and Enhances Antigen Presentation in B-Cell Lymphoma. Blood 2019;134:2858. [Crossref]
- Szydłowski M, Dębek S, Prochorec-Sobieszek M, et al. PIM Kinases Promote Survival and Immune Escape in Primary Mediastinal Large B-Cell Lymphoma through Modulation of JAK-STAT and NF-κB Activity. Am J Pathol 2021;191:567-74. [Crossref] [PubMed]
- Arthur SE, Jiang A, Grande BM, et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat Commun 2018;9:4001. [Crossref] [PubMed]
- Morin RD, Assouline S, Alcaide M, et al. Genetic Landscapes of Relapsed and Refractory Diffuse Large B-Cell Lymphomas. Clin Cancer Res 2016;22:2290-300. [Crossref] [PubMed]
- Bruno A, Alentorn A, Daniau M, et al. TERT promoter mutations in primary central nervous system lymphoma are associated with spatial distribution in the splenium. Acta Neuropathol 2015;130:439-40. [Crossref] [PubMed]
- Montesinos-Rongen M, Schmitz R, Brunn A, et al. Mutations of CARD11 but not TNFAIP3 may activate the NF-kappaB pathway in primary CNS lymphoma. Acta Neuropathol 2010;120:529-35. [Crossref] [PubMed]
- Krumbholz M, Theil D, Derfuss T, et al. BAFF is produced by astrocytes and up-regulated in multiple sclerosis lesions and primary central nervous system lymphoma. J Exp Med 2005;201:195-200. [Crossref] [PubMed]
- Birnbaum T, Langer S, Roeber S, et al. Expression of B-cell activating factor, a proliferating inducing ligand and its receptors in primary central nervous system lymphoma. Neurol Int 2013;5:e4. [Crossref] [PubMed]
- Zhou X, Mulazzani M, von Mücke-Heim IA, et al. The Role of BAFF-R Signaling in the Growth of Primary Central Nervous System Lymphoma. Front Oncol 2020;10:682. [Crossref] [PubMed]
- Thaler FS, Laurent SA, Huber M, et al. Soluble TACI and soluble BCMA as biomarkers in primary central nervous system lymphoma. Neuro Oncol 2017;19:1618-27. [Crossref] [PubMed]
- Mizutani H, Nakane S, Ikeda T, et al. CSF TACI and BAFF levels in patients with primary CNS lymphoma as novel diagnostic biomarkers. Ann Clin Transl Neurol 2018;5:1611-6. [Crossref] [PubMed]
- Mulazzani M, Huber M, Borchard S, et al. APRIL and BAFF: novel biomarkers for central nervous system lymphoma. J Hematol Oncol 2019;12:102. [Crossref] [PubMed]
- Muta H, Sugita Y, Furuta T, et al. Expression of the ghrelin/growth hormone secretagogue receptor axis and its functional role in promoting tumor growth in primary central nervous system lymphomas. Neuropathology 2020;40:232-9. [Crossref] [PubMed]
- Chen JH, Huang SM, Chen CC, et al. Ghrelin induces cell migration through GHS-R, CaMKII, AMPK, and NF-κB signaling pathway in glioma cells. J Cell Biochem 2011;112:2931-41. [Crossref] [PubMed]
- Salmaggi A, Eoli M, Corsini E, et al. Cerebrospinal fluid interleukin-10 levels in primary central nervous system lymphoma: a possible marker of response to treatment? Ann Neurol 2000;47:137-8. [Crossref] [PubMed]
- Sasayama T, Nakamizo S, Nishihara M, et al. Cerebrospinal fluid interleukin-10 is a potentially useful biomarker in immunocompetent primary central nervous system lymphoma (PCNSL). Neuro Oncol 2012;14:368-80. [Crossref] [PubMed]
- Rubenstein JL, Wong VS, Kadoch C, et al. CXCL13 plus interleukin 10 is highly specific for the diagnosis of CNS lymphoma. Blood 2013;121:4740-8. [Crossref] [PubMed]
- Mizowaki T, Sasayama T, Tanaka K, et al. STAT3 activation is associated with cerebrospinal fluid interleukin-10 (IL-10) in primary central nervous system diffuse large B cell lymphoma. J Neurooncol 2015;124:165-74. [Crossref] [PubMed]
- Sasagawa Y, Akai T, Tachibana O, et al. Diagnostic value of interleukin-10 in cerebrospinal fluid for diffuse large B-cell lymphoma of the central nervous system. J Neurooncol 2015;121:177-83. [Crossref] [PubMed]
- Nguyen-Them L, Costopoulos M, Tanguy ML, et al. The CSF IL-10 concentration is an effective diagnostic marker in immunocompetent primary CNS lymphoma and a potential prognostic biomarker in treatment-responsive patients. Eur J Cancer 2016;61:69-76. [Crossref] [PubMed]
- Song Y, Zhang W, Zhang L, et al. Cerebrospinal Fluid IL-10 and IL-10/IL-6 as Accurate Diagnostic Biomarkers for Primary Central Nervous System Large B-cell Lymphoma. Sci Rep 2016;6:38671. [Crossref] [PubMed]
- Lemma SA, Pasanen AK, Haapasaari KM, et al. Similar chemokine receptor profiles in lymphomas with central nervous system involvement - possible biomarkers for patient selection for central nervous system prophylaxis, a retrospective study. Eur J Haematol 2016;96:492-501. [Crossref] [PubMed]
- Rubenstein JL, Geng H, Fraser EJ, et al. Phase 1 investigation of lenalidomide/rituximab plus outcomes of lenalidomide maintenance in relapsed CNS lymphoma. Blood Adv 2018;2:1595-607. [Crossref] [PubMed]
- Costopoulos M, Kim R, Choquet S, et al. Cerebrospinal fluid interleukin (IL)-10 and IL-10:IL-6 ratio as biomarkers for small B-cell lymphoproliferations with leptomeningeal dissemination. Semin Hematol 2018;55:179-81. [Crossref] [PubMed]
- Mondello P, Cuzzocrea S, Arrigo C, et al. STAT6 activation correlates with cerebrospinal fluid IL-4 and IL-10 and poor prognosis in primary central nervous system lymphoma. Hematol Oncol 2020;38:106-10. [Crossref] [PubMed]
- Shao J, Chen K, Li Q, et al. High Level of IL-10 in Cerebrospinal Fluid is Specific for Diagnosis of Primary Central Nervous System Lymphoma. Cancer Manag Res 2020;12:6261-8. [Crossref] [PubMed]
- Tsang M, Cleveland J, Rubenstein JL. On point in primary CNS lymphoma. Hematol Oncol 2020;38:640-7. [Crossref] [PubMed]
- Geng M, Song Y, Xiao H, et al. Clinical significance of interleukin-10 concentration in the cerebrospinal fluid of patients with primary central nervous system lymphoma. Oncol Lett 2021;21:2. [PubMed]
- Fischer L, Korfel A, Pfeiffer S, et al. CXCL13 and CXCL12 in central nervous system lymphoma patients. Clin Cancer Res 2009;15:5968-73. [Crossref] [PubMed]
- Ferreri AJM, Calimeri T, Lopedote P, et al. MYD88 L265P mutation and interleukin-10 detection in cerebrospinal fluid are highly specific discriminating markers in patients with primary central nervous system lymphoma: results from a prospective study. Br J Haematol 2021;193:497-505. [Crossref] [PubMed]
- Gonzalez-Gomez P, Bello MJ, Arjona D, et al. CpG island methylation of tumor-related genes in three primary central nervous system lymphomas in immunocompetent patients. Cancer Genet Cytogenet 2003;142:21-4. [Crossref] [PubMed]
- Venturutti L, Teater M, Zhai A, et al. TBL1XR1 Mutations Drive Extranodal Lymphoma by Inducing a Pro-tumorigenic Memory Fate. Cell 2020;182:297-316.e27. [Crossref] [PubMed]
- Venturutti L, Melnick AM. The dangers of déjà vu: memory B cells as the cells of origin of ABC-DLBCLs. Blood 2020;136:2263-74. [Crossref] [PubMed]
- Pellissery S, Richter J, Haake A, et al. Somatic mutations altering Tyr641 of EZH2 are rare in primary central nervous system lymphoma. Leuk Lymphoma 2010;51:2135-6. [Crossref] [PubMed]
- Lauw MIS, Lucas CG, Ohgami RS, et al. Primary Central Nervous System Lymphomas: A Diagnostic Overview of Key Histomorphologic, Immunophenotypic, and Genetic Features. Diagnostics (Basel) 2020;10:1076. [Crossref] [PubMed]
- Hattab EM, Martin SE, Al-Khatib SM, et al. Most primary central nervous system diffuse large B-cell lymphomas occurring in immunocompetent individuals belong to the nongerminal center subtype: a retrospective analysis of 31 cases. Mod Pathol 2010;23:235-43. [Crossref] [PubMed]
- Pina-Oviedo S, Bellamy WT, Gokden M. Analysis of primary central nervous system large B-cell lymphoma in the era of high-grade B-cell lymphoma: Detection of two cases with MYC and BCL6 rearrangements in a cohort of 12 cases. Ann Diagn Pathol 2020;48:151610. [Crossref] [PubMed]
- Brunn A, Nagel I, Montesinos-Rongen M, et al. Frequent triple-hit expression of MYC, BCL2, and BCL6 in primary lymphoma of the central nervous system and absence of a favorable MYC(low)BCL2 (low) subgroup may underlie the inferior prognosis as compared to systemic diffuse large B cell lymphomas. Acta Neuropathol 2013;126:603-5. [Crossref] [PubMed]
- Nosrati A, Monabati A, Sadeghipour A, et al. MYC, BCL2, and BCL6 rearrangements in primary central nervous system lymphoma of large B cell type. Ann Hematol 2019;98:169-73. [Crossref] [PubMed]
- Fangazio M, Ladewig E, Gomez K, et al. Genetic mechanisms of HLA-I loss and immune escape in diffuse large B cell lymphoma. Proc Natl Acad Sci U S A 2021;118:e2104504118. [Crossref] [PubMed]
- Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet 2011;43:830-7. [Crossref] [PubMed]
- Sethi TK, Kovach AE, Grover NS, et al. Clinicopathologic correlates of MYD88 L265P mutation and programmed cell death (PD-1) pathway in primary central nervous system lymphoma. Leuk Lymphoma 2019;60:2880-9. [Crossref] [PubMed]
- Waldera-Lupa DM, Etemad-Parishanzadeh O, Brocksieper M, et al. Proteomic changes in cerebrospinal fluid from primary central nervous system lymphoma patients are associated with protein ectodomain shedding. Oncotarget 2017;8:110118-32. [Crossref] [PubMed]
- Fischer L, Hummel M, Korfel A, et al. Differential micro-RNA expression in primary CNS and nodal diffuse large B-cell lymphomas. Neuro Oncol 2011;13:1090-8. [Crossref] [PubMed]
- Robertus JL, Harms G, Blokzijl T, et al. Specific expression of miR-17-5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Mod Pathol 2009;22:547-55. [Crossref] [PubMed]
- Zajdel M, Rymkiewicz G, Sromek M, et al. Tumor and Cerebrospinal Fluid microRNAs in Primary Central Nervous System Lymphomas. Cancers (Basel) 2019;11:1647. [Crossref] [PubMed]
- Takashima Y, Kawaguchi A, Iwadate Y, et al. miR-101, miR-548b, miR-554, and miR-1202 are reliable prognosis predictors of the miRNAs associated with cancer immunity in primary central nervous system lymphoma. PLoS One 2020;15:e0229577. [Crossref] [PubMed]
- Roth P, Keller A, Hoheisel JD, et al. Differentially regulated miRNAs as prognostic biomarkers in the blood of primary CNS lymphoma patients. Eur J Cancer 2015;51:382-90. [Crossref] [PubMed]
- Mao X, Sun Y, Tang J. Serum miR-21 is a diagnostic and prognostic marker of primary central nervous system lymphoma. Neurol Sci 2014;35:233-8. [Crossref] [PubMed]
- Baraniskin A, Kuhnhenn J, Schlegel U, et al. MicroRNAs in cerebrospinal fluid as biomarker for disease course monitoring in primary central nervous system lymphoma. J Neurooncol 2012;109:239-44. [Crossref] [PubMed]
- Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 2011;117:3140-6. [Crossref] [PubMed]
- Baraniskin A, Chomiak M, Ahle G, et al. MicroRNA-30c as a novel diagnostic biomarker for primary and secondary B-cell lymphoma of the CNS. J Neurooncol 2018;137:463-8. [Crossref] [PubMed]
- Takashima Y, Kawaguchi A, Iwadate Y, et al. MicroRNA signature constituted of miR-30d, miR-93, and miR-181b is a promising prognostic marker in primary central nervous system lymphoma. PLoS One 2019;14:e0210400. [Crossref] [PubMed]
- Yu X, Li Z, Shen J, et al. Role of microRNAs in primary central nervous system lymphomas. Cell Prolif 2016;49:147-53. [Crossref] [PubMed]
- Baraniskin A, Zaslavska E, Nöpel-Dünnebacke S, et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for primary central nervous system lymphoma. Neuro Oncol 2016;18:361-7. [Crossref] [PubMed]
- Takano S, Hattori K, Ishikawa E, et al. MyD88 Mutation in Elderly Predicts Poor Prognosis in Primary Central Nervous System Lymphoma: Multi-Institutional Analysis. World Neurosurg 2018;112:e69-73. [Crossref] [PubMed]
- Rossi D, Spina V, Bruscaggin A, et al. Liquid biopsy in lymphoma. Haematologica 2019;104:648-52. [Crossref] [PubMed]
- Lakhotia R, Roschewski M. Circulating tumour DNA in B-cell lymphomas: current state and future prospects. Br J Haematol 2021;193:867-81. [Crossref] [PubMed]
- Melani C, Wilson WH, Roschewski M. Liquid biopsy in non-Hodgkin's lymphoma. Hematol Oncol 2019;37:70-4. [Crossref] [PubMed]
- Heger JM, Gödel P, Balke-Want H, et al. Liquid-biopsy based genotyping of primary central nervous system lymphoma (PCNSL). Hematol Oncol 2021;39: [Crossref]
- Mutter JA, Alig S, Lauer EM, et al. Profiling of Circulating Tumor DNA for Noninvasive Disease Detection, Risk Stratification, and MRD Monitoring in Patients with CNS Lymphoma. Blood 2021;138:6. [Crossref]
- Hiemcke-Jiwa LS, Leguit RJ, Snijders TJ, et al. Molecular analysis in liquid biopsies for diagnostics of primary central nervous system lymphoma: Review of literature and future opportunities. Crit Rev Oncol Hematol 2018;127:56-65. [Crossref] [PubMed]
- Bobillo S, Crespo M, Escudero L, et al. Cell free circulating tumor DNA in cerebrospinal fluid detects and monitors central nervous system involvement of B-cell lymphomas. Haematologica 2021;106:513-21. [Crossref] [PubMed]
- Grommes C, Gavrilovic I, Miller AM, et al. Phase Ib of Copanlisib in Combination with Ibrutinib in Recurrent/Refractory Primary CNS Lymphoma (PCNSL). Blood 2019;134:1598. [Crossref]
- Hickmann AK, Frick M, Hadaschik D, et al. Molecular tumor analysis and liquid biopsy: a feasibility investigation analyzing circulating tumor DNA in patients with central nervous system lymphomas. BMC Cancer 2019;19:192. [Crossref] [PubMed]
- Rimelen V, Ahle G, Pencreach E, et al. Tumor cell-free DNA detection in CSF for primary CNS lymphoma diagnosis. Acta Neuropathol Commun 2019;7:43. [Crossref] [PubMed]
- Mendez JS, Grommes C. Treatment of Primary Central Nervous System Lymphoma: From Chemotherapy to Small Molecules. Am Soc Clin Oncol Educ Book 2018;38:604-15. [Crossref] [PubMed]
- Mondello P, Mian M, Bertoni F. Primary central nervous system lymphoma: Novel precision therapies. Crit Rev Oncol Hematol 2019;141:139-45. [Crossref] [PubMed]
- Yang H, Xun Y, Yang A, et al. Advances and challenges in the treatment of primary central nervous system lymphoma. J Cell Physiol 2020;235:9143-65. [Crossref] [PubMed]
- Royer-Perron L, Hoang-Xuan K, Alentorn A. Primary central nervous system lymphoma: time for diagnostic biomarkers and biotherapies? Curr Opin Neurol 2017;30:669-76. [Crossref] [PubMed]
- Cai Q, Fang Y, Young KH. Primary Central Nervous System Lymphoma: Molecular Pathogenesis and Advances in Treatment. Transl Oncol 2019;12:523-38. [Crossref] [PubMed]
- Chihara D, Dunleavy K. Primary Central Nervous System Lymphoma: Evolving Biologic Insights and Recent Therapeutic Advances. Clin Lymphoma Myeloma Leuk 2021;21:73-9. [Crossref] [PubMed]
- Illerhaus G, Schorb E, Kasenda B. Novel agents for primary central nervous system lymphoma: evidence and perspectives. Blood 2018;132:681-8. [Crossref] [PubMed]
- Löw S, Han CH, Batchelor TT. Primary central nervous system lymphoma. Ther Adv Neurol Disord 2018;11:1756286418793562. [Crossref] [PubMed]
- Grommes C, Tang SS, Wolfe J, et al. Phase 1b trial of an ibrutinib-based combination therapy in recurrent/refractory CNS lymphoma. Blood 2019;133:436-45. [Crossref] [PubMed]
- Lionakis MS, Dunleavy K, Roschewski M, et al. Inhibition of B Cell Receptor Signaling by Ibrutinib in Primary CNS Lymphoma. Cancer Cell 2017;31:833-843.e5. [Crossref] [PubMed]
- Grommes C, Piotrowski A, Pentsova E, et al. Phase Ib Trial with Dose Expansion of the Bruton's Tyrosine Kinase (BTK) Inhibitor, Ibrutinib, in Combination with Rituximab and Lenalidomide in Patients with Refractory/Recurrent Primary Central Nervous System Lymphoma (PCNSL) and Refractory/Recurrent Secondary Central Nervous System Lymphoma (SCNSL). Blood 2020;136:48. [Crossref]
- Soussain C, Choquet S, Blonski M, et al. Ibrutinib monotherapy for relapse or refractory primary CNS lymphoma and primary vitreoretinal lymphoma: Final analysis of the phase II 'proof-of-concept' iLOC study by the Lymphoma study association (LYSA) and the French oculo-cerebral lymphoma (LOC) network. Eur J Cancer 2019;117:121-30. [Crossref] [PubMed]
- Korfel A, Schlegel U, Herrlinger U, et al. Phase II Trial of Temsirolimus for Relapsed/Refractory Primary CNS Lymphoma. J Clin Oncol 2016;34:1757-63. [Crossref] [PubMed]
- Tun HW, Johnston PB, DeAngelis LM, et al. Phase 1 study of pomalidomide and dexamethasone for relapsed/refractory primary CNS or vitreoretinal lymphoma. Blood 2018;132:2240-8. [Crossref] [PubMed]
- Ghesquieres H, Chevrier M, Laadhari M, et al. Lenalidomide in combination with intravenous rituximab (REVRI) in relapsed/refractory primary CNS lymphoma or primary intraocular lymphoma: a multicenter prospective 'proof of concept' phase II study of the French Oculo-Cerebral lymphoma (LOC) Network and the Lymphoma Study Association (LYSA)†. Ann Oncol 2019;30:621-8. [Crossref] [PubMed]
- Grommes C, Pentsova E, Nolan C, et al. Phase II study of single agent buparlisib in recurrent/refractory primary (PCNSL) and secondary CNS lymphoma (SCNSL). Ann Oncol 2016;27:VI106. [Crossref]
- Nowakowski GS, Leslie LA, Joffe E, et al. A Multi-Center, Dose-Finding Study to Assess Safety, Tolerability, Pharmacokinetics and Preliminary Efficacy of a Novel IRAK4 Inhibitor CA-4948 in Combination with Ibrutinib, in Patients with Relapsed or Refractory Hematologic Malignancies. Blood 2020;136:49-50. [Crossref]
- Foster J, Muscal JA, Minard CG, et al. Phase 1 study of pevonedistat (MLN4924) in combination with temozolomide (TMZ) and irinotecan (IRN) in pediatric patients with recurrent or refractory solid tumors (ADVL1615). J Clin Oncol 2019;37:e21521. [Crossref]
- Westin JR, Fowler NH, Nastoupil LJ, et al. A Phase II Trial of Nivolumab and Ibrutinib for Patients with Relapsed or Refractory Central Nervous System Lymphoma. Blood 2019;134:4086. [Crossref]
- Roschewski M, Melani C, Lakhotia R, et al. Phase 1 Study of Escalating Doses of Ibrutinib and Temozolomide, Etoposide, Liposomal Doxorubicin, Dexamethasone, Rituximab (TEDDI-R) with Isavuconazole for Relapsed and Refractory Primary CNS Lymphoma. Blood 2020;136:12-3. [Crossref]
- Chamoun K, Choquet S, Boyle E, et al. Ibrutinib monotherapy in relapsed/refractory CNS lymphoma: A retrospective case series. Neurology 2017;88:101-2. [Crossref] [PubMed]
- Phelan JD, Young RM, Webster DE, et al. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature 2018;560:387-91. [Crossref] [PubMed]
- Goldwirt L, Beccaria K, Ple A, et al. Ibrutinib brain distribution: a preclinical study. Cancer Chemother Pharmacol 2018;81:783-9. [Crossref] [PubMed]
- Pouzoulet F, Rezai K, Li Z, et al. Preclinical Evaluation of Ibrutinib for Central Nervous System Lymphoma. Blood 2016;128:4170. [Crossref]
- Takashima Y, Hayano A, Yamanaka R. Metabolome Analysis Reveals Excessive Glycolysis via PI3K/AKT/mTOR and RAS/MAPK Signaling in Methotrexate-Resistant Primary CNS Lymphoma-Derived Cells. Clin Cancer Res 2020;26:2754-66. [Crossref] [PubMed]
- Nitta N, Nakasu S, Shima A, et al. mTORC1 signaling in primary central nervous system lymphoma. Surg Neurol Int 2016;7:S475-80. [Crossref] [PubMed]
- Marosvári D, Nagy N, Kriston C, et al. Discrepancy Between Low Levels of mTOR Activity and High Levels of P-S6 in Primary Central Nervous System Lymphoma May Be Explained by PAS Domain-Containing Serine/Threonine-Protein Kinase-Mediated Phosphorylation. J Neuropathol Exp Neurol 2018;77:268-73. [Crossref] [PubMed]
- Yang Y, Shaffer AL 3rd, Emre NC, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell 2012;21:723-37. [Crossref] [PubMed]
- Jiménez I, Carabia J, Bobillo S, et al. Repolarization of tumor infiltrating macrophages and increased survival in mouse primary CNS lymphomas after XPO1 and BTK inhibition. J Neurooncol 2020;149:13-25. [Crossref] [PubMed]
- Cani AK, Hovelson DH, Demirci H, et al. Next generation sequencing of vitreoretinal lymphomas from small-volume intraocular liquid biopsies: new routes to targeted therapies. Oncotarget 2017;8:7989-98. [Crossref] [PubMed]
- Marjon K, Cameron MJ, Quang P, et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep 2016;15:574-87. [Crossref] [PubMed]
- Bewarder M, Kiefer M, Moelle C, et al. Integration of the B-Cell Receptor Antigen Neurabin-I/SAMD14 Into an Antibody Format as New Therapeutic Approach for the Treatment of Primary CNS Lymphoma. Front Oncol 2020;10:580364. [Crossref] [PubMed]
- Bewarder M, Körbel C, Laschke MW, et al. The B-Cell Receptor Antigen ARS2 Can be Integrated into a BAR-Body Format to Treat Diffuse Large B-Cell Lymphomas in Xenograft Mouse Models. Blood 2019;134:2860. [Crossref]
- Spies E, Fichtner M, Müller F, et al. Comment on “Primary Central Nervous System (CNS) Lymphoma B Cell Receptors Recognize CNS Proteins”. J Immunol 2015;195:4549-50. [Crossref] [PubMed]
- Kurzwelly D, Glas M, Roth P, et al. Primary CNS lymphoma in the elderly: temozolomide therapy and MGMT status. J Neurooncol 2010;97:389-92. [Crossref] [PubMed]
- Adachi J, Mishima K, Wakiya K, et al. O6-methylguanine-DNA methyltransferase promoter methylation in 45 primary central nervous system lymphomas: quantitative assessment of methylation and response to temozolomide treatment. J Neurooncol 2012;107:147-53. [Crossref] [PubMed]
- Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997-1003. [Crossref] [PubMed]
- Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000;343:1350-4. [Crossref] [PubMed]
- Jiang X, Reardon DA, Desjardins A, et al. O6-methylguanine-DNA methyltransferase (MGMT) immunohistochemistry as a predictor of resistance to temozolomide in primary CNS lymphoma. J Neurooncol 2013;114:135-40. [Crossref] [PubMed]
- Ferreri AJ, Dell'Oro S, Capello D, et al. Aberrant methylation in the promoter region of the reduced folate carrier gene is a potential mechanism of resistance to methotrexate in primary central nervous system lymphomas. Br J Haematol 2004;126:657-64. [Crossref] [PubMed]
- Karschnia P, Blobner J, Teske N, et al. CAR T-Cells for CNS Lymphoma: Driving into New Terrain? Cancers (Basel) 2021;13:2503. [Crossref] [PubMed]
- Qualls D, Salles G. Optimizing CAR T cell therapy in lymphoma. Hematol Oncol 2021;39:104-12. [Crossref] [PubMed]
- Alcantara M, Houillier C, Le Garff-Tavernier M, et al. CAR-T cell therapy in primary central nervous system lymphoma (PCNSL): the experience of the French network for oculo-cerebral lymphomas (LOC). Hematol Oncol 2021;39: [Crossref]
- Brunn A, Montesinos-Rongen M, Strack A, et al. Expression pattern and cellular sources of chemokines in primary central nervous system lymphoma. Acta Neuropathol 2007;114:271-6. [Crossref] [PubMed]
- Poulain S, Boyle EM, Tricot S, et al. Absence of CXCR4 mutations but high incidence of double mutant in CD79A/B and MYD88 in primary central nervous system lymphoma. Br J Haematol 2015;170:285-7. [Crossref] [PubMed]
- Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol 2011;11:597-606. [Crossref] [PubMed]
Cite this article as: Bertoni F, Montesinos-Rongen M. Primary diffuse large B-cell lymphoma of the central nervous system: molecular and biological features of neoplastic cells. Ann Lymphoma 2022;6:1.


