
Adult T-cell leukemia/lymphoma—pathobiology and implications for modern clinical management
Introduction
Adult T-cell leukemia/lymphoma (ATL) is an aggressive CD4+ T-cell malignancy that arises in 2–5% of individuals chronically infected with human T-cell lymphotropic virus type 1 (HTLV-1). ATL presents with diverse clinical features, including circulating leukemic cells, generalized lymph node swelling, hepatosplenomegaly, skin involvement, opportunistic infections, and hypercalcemia and has a poor prognosis, with shorter overall survival relative to other peripheral T-cell lymphomas. Although progress has been made in understanding the biology that underpins ATL, treatment remains unsatisfactory, particularly for those with primary refractory or relapsed refractory disease.
Which HTLV-1 carriers develop ATL?
Chronic infection with the deltaretrovirus HTLV-1 is the only cause of ATL, which arises decades after infection, arising most commonly in carriers infected during infancy through breastfeeding. The lifetime risk of ATL among HTLV-1 carriers has been estimated to be ~2.5–6% in several endemic areas of Japan (1-3). Globally, there are an estimated 5–10 million people infected with HTLV-1, although this is likely to be a conservative estimate since it is based on epidemiological studies in countries that screen blood donors, which are often low prevalence nations (4). Although it is a blood borne virus, in the UK there is no systematic screening for HTLV-1 infection in antenatal or genitourinary clinics, but testing is undertaken in first-time blood donors, and donors and recipients of stem cell or solid organ transplants.
HTLV-1 is endemic in Japan, the Caribbean, Central and South America, parts of sub-Saharan Africa, Middle East, Melanesia, central Australia and Romania. In the USA and Europe, most prevalent in immigrants from endemic regions and their descendants. The geographic presentation of ATL corresponds with the known epidemiology of HTLV-1. For example, ATL accounts for approximately 25% of peripheral T-cell lymphomas in Asia (primarily Japan), compared with 2% in North America and 1% in Europe (5). The age at onset varies by region. The median age at presentation in Caribbean and South America is 40–50 years. By contrast, in Japan the median age at presentation has been recently reported at 67.5 years which has risen from 56.9 years in 1984–1985, and 58.9 years in 1992–1993 (6). This progressive rise in the age at presentation suggests a cohort effect, i.e., a cohort of the population in Japan that developed a high prevalence of HTLV-1 infection in the 1920s and 1930s. In Japan ATL occurs more frequently in men, although the sex ratio is falling (6), and this male predominance is not observed in Jamaica (7). By contrast a study from Brooklyn, USA suggested a clear female predominance (8).
HTLV-1 infection alone is not sufficient to drive the development of ATL, and the majority of carriers will remain asymptomatic for life. Reported risk factors include ageing (9), a family history of ATL or HTLV-1 associated myelopathy (HAM) (9), an immunocompromised clinical status, and a proviral load (PVL) >4% [PVL, proportion of infected peripheral blood mononuclear cells (PBMCs)]. A higher PVL is an independent risk factor for ATL, even after adjusting for sex, age, and family history (10), and the risk of ATL is carried amongst high load carriers whose individual lifetime risk is >20% (11,12).
Diagnosis & prognosis
Diagnosis of ATL is determined by the combination of clinical presentation (typically leukocytosis, lymphadenopathy, hypercalcaemia, lytic bone lesions, skin lesions, raised LDH, often with opportunistic infections), a characteristic blood film morphology, and immunophenotyping of blood or tissue biopsy. The malignant T-cells typically demonstrate polylobated nuclei (“flower cells”) with condensed chromatin, and agranular cytoplasm and express CD3dim+ CD4+ CD5-CD7- CD25+ CCR4+ and a monoclonal TCRVβ. Some cases may be CD8+ CD4-, CD4+CD8+, or CD8-CD4-. The criteria for ATL diagnosis and subtype classification remain as defined by Shimoyama et al. (13,14) (summarised in Table 1). Several factors that predict poor prognosis include a poor performance status, elevated LDH level, hypercalcaemia, nodal sites >4, age >40 years, thrombocytopenia, eosinophilia, bone marrow involvement, high serum level of interleukin 5, C-C chemokine receptor 4 expression (CCR4), lung resistance-related protein, p53 mutation, and p16 deletion (14), and there is evidence of a higher frequency of these poor prognostic risk factors among Caribbean patients than in those from Japan (15). Depressingly, the median overall survival of ATL in Japanese patients treated with chemotherapy has been largely unchanged in the nearly 40 years since the syndrome was first described (13,16). Risk models that have incorporated combinations of these prognostic risk factors have been developed, and these models may in future guide chemotherapy or targeted agent treatment selection (17,18). Currently the median survival of those with acute or lymphoma ATL remains 9–13 months, and those with chronic or smouldering ATL 2–4 years (16). Death frequently occurs from treatment-refractory disease, transformation from indolent to aggressive subtypes, or opportunistic infection.
Table 1
| Subtype | Asymptomatic HTLV- 1 carrier | Smouldering | Chronic | Acute | Lymphoma |
|---|---|---|---|---|---|
| HTLV-1 serology | + | + | + | + | + |
| Monoclonal integration of provirus | – | + | + | + | + (lymph node) |
| Lymphocyte count | Normal | Normal | Elevated | Elevated | Normal |
| Percentage circulating abnormal lymphocytes | <5% | ≥5% Or <5% if ATL skin & lung lesions present | ≥5% | ≥5% | ≤1% |
| Hypercalcaemia | – | – | – | Normal or high | Normal or high |
| LDH | Normal | Normal or ≤1.5 ULN | Normal or <2 ULN | Normal or high | Normal or high |
| Lymphadenopathy | – | – | +/– | +/– | + |
| Skin or lung involvement | – | +/– | +/– | +/– | +/– |
| Bone marrow or splenic involvement | – | – | +/– | +/– | +/– |
| Bone, gastrointestinal or central nervous system involvement | – | – | – | +/– | +/– |
ATL, adult T-cell leukemia/lymphoma; ULN, upper limit of normal.
Treatment strategies for ATL are based primarily on the disease subtype. Current frontline treatment options for ATL vary by geographic location and include active monitoring (in Japan for indolent subtypes), zidovudine plus interferon-alfa (ZDV/IFN) where available, multiagent chemotherapy, and for a subset of suitable patients allogeneic hematopoietic stem cell transplantation. Novel agents with approvals in Japan include mogamulizumab (a monoclonal antibody against the chemokine receptor CCR4) and lenalidomide. Current treatment strategies are reviewed elsewhere (19,20).
Viral determinants in the development ATL
HTLV-1-infected T-cell clones appear to persist indefinitely during chronic infection. The clones replicate more frequently than uninfected T-cells (21) and as a consequence replicative errors are likely to make a significant contribution to genomic instability and cellular transformation. Like other retroviruses the integrated HTLV-1 proviral genome consists of two long terminal repeats, structural and regulatory genes and an additional sequence designated pX, which encodes proteins including two crucial proteins that regulate the persistence, expression and pathogenesis of HTLV-1: Tax and HBZ (HTLV-1 B-zip protein). Tax is a transcriptional transactivator protein which, in addition to driving replication of HTLV-1, strongly activates cellular gene expression in pathways involved in the proliferation of T-lymphocytes, mainly via the activation of NF-κB and AP-1 pathways. Tax-expressing cells may bypass normal cell-cycle checkpoints and affect DNA damage and apoptosis pathways, resulting in the accumulation of genetic, epigenetic and RNA modifications. Tax-transgenic mice develop tumours including T-cell lymphoma, and so Tax has been considered to be a critical viral factor involved in the immortalization and transformation of infected cells. HBZ messenger RNA was also shown to also promote proliferation of ATL cells and has a variety of functions, including modulation of Tax expression. HBZ also drives cell proliferation, increases hTERT transcription, inhibits apoptosis and interferes with micro RNAs and HBZ transgenic mice also develop T-cell lymphomas as well as systemic inflammation (reviewed (22-24). However, neither tax nor HBZ should be considered as oncogenes, such as those of acutely transforming retroviruses e.g., Rous sarcoma virus containing viral-src, but that oncogenesis is an indirect, and fortunately, uncommon consequence of the persistent replication of HTLV-1 infected cells, which in turn depend upon the cooperative expression and actions of these two genes.
Expression of the HTLV-1 proteins in turn exposes infected cells to virus-specific cytotoxic T-lymphocytes (CTL), and the efficiency or ‘quality’ of the CTL response is a key determinant in the host PVL, and the likely clinical outcome of infection (25). The key factor that determines a high quality anti-HTLV-1 CTL response are the host genotype in the HLA Class 1 and killer immunoglobulin-like receptor (KIR) loci, and the ability to present epitopes from the HBZ antigen to CTLs (22,25). Thus, virus-infected CD4+ T cell clones continue to proliferate over decades of infection, with selection of clones carrying mutations that promote T-lymphocyte proliferation and survival, and those whose pattern of intermittent HTLV-1 reactivation minimizes their exposure to the CTL response. CTL escape is evident in ATL: immunodominant viral epitopes and host genes (e.g., HLA-A and HLA-B) involved in antigen presentation are frequently silenced and/or deleted, and genes implicated in reduced immune surveillance (PD-L1) are overexpressed (26-28).
The role of the stromal microenvironment—particularly fibroblasts—on immortalisation, tumour induction and propagation has been extensively reported in epithelial cancers, but is less well understood in HTLV-1 infection and ATL. There are three main reasons for this. First, ex vivo culture rapidly alters HTLV-1 expression, stimulating a burst of transcription of the proviral plus strand. Second, there is a lack of robust animal models of HTLV-1 transformation. Third, HTLV-1 infection typically remains clinically latent for decades before the presentation of ATL. There have been reports that co-culture of murine marrow stromal cells with either ex vivo ATL cells, HTLV-1-infected cells or ATL cell lines, can support the growth of primary cells and cell lines (29); however, HTLV-1 does not infect mouse cells, and the physiological significance of these findings is unclear. ATL cell lines and primary cells, co-cultured with epithelial like cells were found to be protected from apoptosis, became quiescent and acquired a cancer stem cell-like phenotype (30). Finally, stromal cells may suppress viral expression via type 1 interferons (31).
Epigenetic modifications in ATL cells may promote both cell survival and chemotherapy resistance. The genome of the ATL clone frequently has extensive CpG island DNA hypermethylation with associated transcriptional silencing. Approximately 40% of ATL samples were associated with a CpG island methylator phenotype (CIMP) and the CIMP status was associated with aggressive clinical ATL phenotypes. However, ATL was not associated with mutations in known epigenetic regulators in other cancers (TET2, IDH2 and DNMT3A). HLA class 1 genes were also either silenced by hypermethylation or lost in many cases of ATL, contributing to immune evasion by ATL cells (26,32). Although abnormalities in histone acetylation have not been well described in ATL, histone deacetylase inhibitors can induce apoptosis and inhibit NF-κB expression in HTLV-1 and ATL cell-lines (33,34). Clinical trials of HDAC inhibitors in T-cell malignancies are ongoing, but future studies to understand the mechanism and efficacy in HTLV-1 and ATL are required. A recent comprehensive study of the polycomb-dependent epigenetic and transcriptomic landscape of ATL cells revealed genome-wide deposition of H3K27me3, the characteristic mark of polycomb repressive complex 2 (PRC2) (35). H3K27me3 was abnormally increased in over half the genes (53.8%) with consequent downregulation of key gene expression (miR-31, BCL2, EVC1/2, CDKN1A and NDRG2) associated with disease progression from indolent to aggressive ATL. An international clinical trial of an oral dual inhibitor of EZH1/2 is underway (clinical trials ID NCT04102150).
Clonality in HTLV-1
A typical healthy carrier with HTLV-1 infection carries tens of thousands of infected T-cell clones, usually with one copy of HTLV-1 integrated in each genome, which can be distinguished by a unique viral integration site (36). Integration occurs throughout the human genome: it is not random but is biased toward transcriptionally active regions and near certain transcription factor binding sites, including those for STAT1, TP53 and HDAC6 (37). Using high throughput sequencing, within an individual, ATL cells are typically characterised by a single dominant integration site (91% cases), and in ~10% of ATL cases, the malignant clone contains two or more copies (38), which is consistent with earlier reports using Southern blot methods (39,40). Integration sites were originally mapped and enumerated by Southern blot analysis, but more recently high-throughput sequencing methods have led to significantly increased estimates of clone frequency and abundance of non-malignant clones (41-43). In ATL cells, there are no hot-spots of integration in the host genome. Integration within 10 kb of a known oncogene confers a survival advantage in vivo, but this does not appear to contribute directly to leukaemogenesis (38). However, integration within 15 kb upstream of host genes that are frequently dysregulated in other leukemias occurs more frequently than expected by chance (accounting for ~6% cases of ATL) (38) and that integration in the vicinity of cancer drivers may be affected either by provirus-dependent transcription termination or as a result of viral antisense RNA-dependent cis-pertubation (44). More recently it was shown that the HTLV-1 provirus has the ability to alter the host chromatin structure and disrupt host gene regulation: CTCF, a master regulator of chromatin structure and gene expression, binds to HTLV-1, forms loops between the provirus and host genome, and alters the expression of proviral and host genes (45).
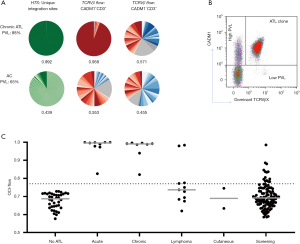
During chronic infection, low-abundance HTLV-1 infected clones make up most of the HTLV-1 PVL. The PVL determines both the risk of ATL and the risk of associated inflammatory diseases, and the PVL correlates with the total number of clones within in an individual and not the degree of oligoclonal proliferation (42). However, when one or more clones have started to undergo malignant transformation, they constitute a disproportionately high fraction of the PVL, even before clinical presentation. In an observational follow up study of HTLV-1 carriers with ‘monoclonal’ populations of HTLV-1 infected cells in the peripheral blood (detected by low resolution, semi quantitative Southern blot), 42% developed ATL in a 20-year period (48 cases/1,000 carrier years). This disproportionate clonal proliferation can now be rigorously quantified by the oligoclonality index (OCI); Figure 1]. The OCI score, derived from the Gini index, was initially applied to precise integration site mapping and clonal abundance following linker mediated PCR and high throughput sequencing. However since this method is technically and bioinfomatically demanding, we developed a flow cytometric assay to estimate the OCI—the ‘OCI-flow’—by quantifying TCRVβ usage in the HTLV-1-infected CADM1+CCR4+CD26- T cells; this method can more easily be applied in diagnostic laboratories (46). An OCI-flow of zero indicates that all TCRVβ subunits have the same frequency in the studied cellular population (i.e., polyclonality), and an OCI-flow of one indicates that only a single clonal population is present (monoclonal), with all cells expressing one TCRVβ subunit. We have recently reported that 19% of high PVL carriers (>4 copies of the HTLV-1 provirus per 100 PBMCs) have PBMCs with oligoclonality scores in the ATL range (OCI-flow >0.770) (47). In samples from these carriers, ATL-like clonal expansions comprised >2% of CD3+ T cells. Two of the asymptomatic carriers with ATL-like clones have since transformed to aggressive ATL, and others had a first-degree relative with ATL (a known risk factor for ATL). These data strongly suggest that these ‘ATL-like’ clones represent premalignant lesions, although full transformation of expanded clones likely requires additional genetic events.
Somatic evolution in ATL
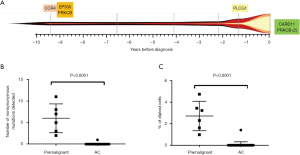
A landmark report from Japan in 2015 described the genomic landscape of ATL and showed a high frequency of specific gain-of-function mutations in ATL, particularly in genes involved in the T-cell receptor and NF-κB signalling pathways, T cell trafficking and immunosurveillance. Frequently mutated genes include PLCG1, PRKCB, CARD11, STAT3, VAV1, NOTCH1, IRF4, CCR4, CCR7, TP53 and CDKN2A (26). These variants were designated potential ATL driver mutations and have since also been observed in an independent cohort of patients with ATL of Afro-Caribbean descent (48).
We hypothesised that ATL-driver mutations are acquired in a stepwise process, and some will be present in the circulation before the diagnosis of ATL. In a cohort of asymptomatic carriers who later developed ATL, we recently reported that mutations accumulate in the ATL-like CADM1+CCR4+CD26- T cells in the circulation in a stepwise manner during the premalignant stage, before the development of aggressive ATL (27). Cells carrying known ATL-driver mutations circulate at high frequencies in the blood (>5% of mononuclear cells) up to 10 years before the diagnosis of ATL, but were not observed in high-PVL asymptomatic HTLV-1 carriers (AC) followed up for a median of 10 years who did not develop ATL
These mutational profiles may also have a future role as disease biomarkers that can be used for diagnosis, prognosis or for tailoring personalised treatment decisions. We have preliminary evidence that lymphoma type ATL-derived cell-free DNA (cfDNA) is released into the plasma and can be used for non-invasive genomic profiling. In normal physiology, short fragments (~145 bp) of degraded genomic DNA are constantly released into extracellular fluids by dying cells. Turnover of HTLV-1-infected cells within tissues releases cfDNA into the plasma. We detected HTLV-1 proviruses in cfDNA extracted from blood plasma from HTLV-1-infected individuals and in patients with lymphoma, the quantity of HTLV-1 proviruses found in cfDNA was several-fold higher than expected. Thus, plasma sampling and genomic analysis of cfDNA may also be used to longitudinally monitor the evolution of lymphoma-type ATL.
Future perspectives
The exciting results from mutational profiling in ATL need to be validated in larger cohorts of patients. But further questions arise: how can we use these tests in practice, at speed and reduced cost? Which mutations are most important? Is the order of acquisition critical to disease development? And, can mutational profiling predict the response to immunotherapy? The combination of PVL measurement, OCI-flow score and targeted sequencing of genomic or cfDNA may make possible a practical prognostic algorithm to identify HTLV-1 carriers at risk of ATL, to select appropriate novel therapies, and to monitor patients with ATL when either on or off therapy.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Christopher P. Fox, Claire Shannon-Lowe) for the series “Lymphoma and Viruses” published in Annals of Lymphoma. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aol-21-6). All authors report grant support from National Institute for Health Biomedical Research Centre. The series “Lymphoma and viruses” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interests to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Arisawa K, Soda M, Endo S, et al. Evaluation of adult T-cell leukemia/lymphoma incidence and its impact on non-Hodgkin lymphoma incidence in southwestern Japan. Int J Cancer 2000;85:319-24. [Crossref] [PubMed]
- Kondo T, Kono H, Miyamoto N, et al. Age- and sex-specific cumulative rate and risk of ATLL for HTLV-I carriers. Int J Cancer 1989;43:1061-4. [Crossref] [PubMed]
- Tokudome S, Tokunaga O, Shimamoto Y, et al. Incidence of adult T-cell leukemia/lymphoma among human T-lymphotropic virus type I carriers in Saga, Japan. Cancer Res 1989;49:226-8. [PubMed]
- Gessain A, Cassar O. Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front Microbiol 2012;3:388. [Crossref] [PubMed]
- Vose J, Armitage J, Weisenburger D. International T-Cell Lymphoma Project. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol 2008;26:4124-30. [Crossref] [PubMed]
- Iwanaga M. Epidemiology of HTLV-1 Infection and ATL in Japan: An Update. Front Microbiol 2020;11:1124. [Crossref] [PubMed]
- Hisada M, Stuver SO, Okayama A, et al. Persistent Paradox of Natural History of Human T Lymphotropic Virus Type I: Parallel Analyses of Japanese and Jamaican Carriers. J Infect Dis 2004;190:1605-9. [Crossref] [PubMed]
- Levine PH, Dosik H, Joseph EM, et al. A study of adult T-cell leukemia/lymphoma incidence in central Brooklyn. Int J Cancer 1999;80:662-6. [Crossref] [PubMed]
- Iwanaga M, Watanabe T, Yamaguchi K. Adult T-cell leukemia: a review of epidemiological evidence. Front Microbiol 2012;3:322. [Crossref] [PubMed]
- Iwanaga M, Watanabe T, Utsunomiya A, et al. Human T-cell leukemia virus type I (HTLV-1) proviral load and disease progression in asymptomatic HTLV-1 carriers: A nationwide prospective study in Japan. Blood 2010;116:1211-9. [Crossref] [PubMed]
- Malik B, Taylor GP. Can we reduce the incidence of adult T-cell leukaemia/lymphoma? Cost-effectiveness of human T-lymphotropic virus type 1 (HTLV-1) antenatal screening in the United Kingdom. Br J Haematol 2019;184:1040-3. [Crossref] [PubMed]
- Demontis MA, Hilburn S, Taylor GP. Human T cell lymphotropic virus type 1 viral load variability and long-term trends in asymptomatic carriers and in patients with human T cell lymphotropic virus type 1-related diseases. AIDS Res Hum Retroviruses 2013;29:359-64. [Crossref] [PubMed]
- Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984-87). Br J Haematol 1991;79:428-37. [Crossref] [PubMed]
- Tsukasaki K, Hermine O, Bazarbachi A, et al. Definition, prognostic factors, treatment, and response criteria of adult T-cell leukemia-lymphoma: a proposal from an international consensus meeting. J Clin Oncol 2009;27:453-9. [Crossref] [PubMed]
- Zell MI, Assal A, Konda B, et al. Analysis of Large Cohort Shows That Caribbean Adult T Cell Leukemia/Lymphoma Is a Chemotherapy Refractory Disease with Very Poor Prognosis That Behaves Distinctly from Japanese Subtypes. Blood 2014;124:1685. [Crossref]
- Katsuya H, Ishitsuka K, Utsunomiya A, et al. Treatment and survival among 1594 patients with ATL. Blood 2015;126:2570-7. [Crossref] [PubMed]
- International Non-Hodgkin's Lymphoma Prognostic Factors Project. A predictive model for aggressive non-Hodgkin's lymphoma. N Engl J Med 1993;329:987-94. [Crossref] [PubMed]
- Fukushima T, Nomura S, Shimoyama M, et al. Japan Clinical Oncology Group (JCOG) prognostic index and characterization of long-term survivors of aggressive adult T-cell leukaemia-lymphoma (JCOG0902A). Br J Haematol 2014;166:739-48. [Crossref] [PubMed]
- Cook LB, Fuji S, Hermine O, et al. Revised adult T-cell leukemia-lymphoma international consensus meeting report. J Clin Oncol 2019;37:677-87. [Crossref] [PubMed]
- Cook LB, Phillips AA. How I treat adult T-cell leukemia/lymphoma. Blood 2021;137:459-70. [Crossref] [PubMed]
- Asquith B, Zhang Y, Mosley AJ, et al. In vivo T lymphocyte dynamics in humans and the impact of human T-lymphotropic virus 1 infection. Proc Natl Acad Sci U S A 2007;104:8035-40. [Crossref] [PubMed]
- Bangham CR, Ratner L. How does HTLV-1 cause adult T-cell leukaemia/lymphoma (ATL)? Curr Opin Virol 2015;14:93-100. [Crossref] [PubMed]
- Nicot C. HTLV-I Tax-Mediated Inactivation of Cell Cycle Checkpoints and DNA Repair Pathways Contribute to Cellular Transformation: “A Random Mutagenesis Model. J Cancer Sci 2015;2: [Crossref] [PubMed]
- Mesnard JM, Barbeau B, Césaire R, Péloponèse JM. Roles of HTLV-1 basic Zip factor (HBZ) in viral chronicity and leukemic transformation. potential new therapeutic approaches to prevent and treat HTLV-1-related diseases. Viruses 2015;7:6490-505. [Crossref] [PubMed]
- Bangham CR. CTL quality and the control of human retroviral infections. Eur J Immunol 2009;39:1700-12. [Crossref] [PubMed]
- Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet 2015;47:1304-15. [Crossref] [PubMed]
- Rowan AG, Dillon R, Witkover A, et al. Evolution of retrovirus-infected premalignant T-cell clones prior to adult T-cell leukemia/lymphoma diagnosis. Blood 2020;135:2023-32. [Crossref] [PubMed]
- Kataoka K, Shiraishi Y, Takeda Y, et al. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature 2016;534:402-6. [Crossref] [PubMed]
- Nagai K, Jinnai I, Hata T, et al. Adhesion-dependent growth of primary adult T cell leukemia cells with down-regulation of HTLV-I p40Tax protein: a novel in vitro model of the growth of acute ATL cells. Int J Hematol 2008;88:551-64. [Crossref] [PubMed]
- Miyatake Y, Sheehy N, Ikeshita S, et al. Anchorage-dependent multicellular aggregate formation induces CD44 high cancer stem cell-like ATL cells in an NF-κB- and vimentin-dependent manner. Cancer Lett 2015;357:355-63. [Crossref] [PubMed]
- Kinpara S, Kijiyama M, Takamori A, et al. Interferon-α (IFN-α) suppresses HTLV-1 gene expression and cell cycling, while IFN-α combined with zidovudine induces p53 signaling and apoptosis in HTLV-1-infected cells. Retrovirology 2013;10:52. [Crossref] [PubMed]
- Watanabe T. Adult T-cell leukemia: molecular basis for clonal expansion and transformation of HTLV-1–infected T cells. Blood 2017;129:1071-81. [Crossref] [PubMed]
- Nishioka C, Ikezoe T, Yang J, et al. Histone deacetylase inhibitors induce growth arrest and apoptosis of HTLV-1-infected T-cells via blockade of signaling by nuclear factor kappaB. Leuk Res 2008;32:287-96. [Crossref] [PubMed]
- Hasegawa H, Yamada Y, Tsukasaki K, et al. LBH589, a deacetylase inhibitor, induces apoptosis in adult T-cell leukemia/lymphoma cells via activation of a novel RAIDD-caspase-2 pathway. Leukemia 2011;25:575-87. [Crossref] [PubMed]
- Fujikawa D, Nakagawa S, Hori M, et al. Polycomb-dependent epigenetic landscape in adult T-cell leukemia. Blood 2016;127:1790-802. [Crossref] [PubMed]
- Cook LB, Rowan AG, Melamed A, et al. HTLV-1-infected T cells contain a single integrated provirus in natural infection. Blood 2012;120:3488-90. [Crossref] [PubMed]
- Melamed A, Laydon DJ, Gillet NA, et al. Genome-wide determinants of proviral targeting, clonal abundance and expression in natural HTLV-1 infection. PLoS Pathog 2013;9:e1003271 [Crossref] [PubMed]
- Cook LB, Melamed A, Niederer H, et al. The role of HTLV-1 clonality, proviral structure, and genomic integration site in adult T-cell leukemia/lymphoma. Blood 2014;123:3925-31. [Crossref] [PubMed]
- Shimamoto Y, Miyahara M, Yamada H, et al. Adult T-cell leukaemia/lymphoma with multiple integrations of human T-cell lymphotropic virus type I proviral DNA: differing clinical features are linked to varied proviral integration. Br J Haematol 1996;92:632-8. [Crossref] [PubMed]
- Tsukasaki K, Tsushima H, Yamamura M, et al. Integration patterns of HTLV-I provirus in relation to the clinical course of ATL: frequent clonal change at crisis from indolent disease. Blood 1997;89:948-56. [Crossref] [PubMed]
- Yamaguchi K, Seiki M, Yoshida M, et al. The detection of human T cell leukemia virus proviral DNA and its application for classification and diagnosis of T cell malignancy. Blood 1984;63:1235-40. [Crossref] [PubMed]
- Gillet NA, Malani N, Melamed A, et al. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood 2011;117:3113-22. [Crossref] [PubMed]
- Katsuya H, Islam S, Tan BJY, et al. The Nature of the HTLV-1 Provirus in Naturally Infected Individuals Analyzed by the Viral DNA-Capture-Seq Approach. Cell Rep 2019;29:724-35.e4. [Crossref] [PubMed]
- Rosewick N, Durkin K, Artesi M, et al. Cis -perturbation of cancer drivers by the HTLV-1/BLV proviruses is an early determinant of leukemogenesis. Nat Commun 2017;8:15264. [Crossref] [PubMed]
- Satou Y, Miyazato P, Ishihara K, et al. The retrovirus HTLV-1 inserts an ectopic CTCF-binding site into the human genome. Proc Natl Acad Sci U S A 2016;113:3054-9. [Crossref] [PubMed]
- Rowan AG, Witkover A, Melamed A, et al. T Cell Receptor Vβ Staining Identifies the Malignant Clone in Adult T cell Leukemia and Reveals Killing of Leukemia Cells by Autologous CD8+ T cells. PLoS Pathog 2016;12:e1006030 [Crossref] [PubMed]
- Wolf SN, Haddow J, Greiller C, et al. Quantification of T cell clonality in human T cell leukaemia virus type-1 carriers can detect the development of adult T cell leukaemia early. Blood Cancer J 2021;11:66. [Crossref] [PubMed]
- Marçais A, Lhermitte L, Artesi M, et al. Targeted deep sequencing reveals clonal and subclonal mutational signatures in Adult T-cell leukemia/lymphoma and defines an unfavorable indolent subtype. Leukemia 2021;35:764-76. [Crossref] [PubMed]
Cite this article as: Cook L, Rowan A, Bangham C. Adult T-cell leukemia/lymphoma—pathobiology and implications for modern clinical management. Ann Lymphoma 2021;5:29.


