
Mantle cell lymphoma pathology update in the 2016 WHO classification
Introduction
Mantle cell lymphoma (MCL) is defined in the WHO classification as a mature B-cell neoplasm usually composed of monomorphic small to medium sized lymphoid cells with irregular nuclei. Centroblasts, paraimmunoblasts and pseudofollicles are absent. The tumor cells are of B-cell phenotype frequently coexpressing CD5 (1). This neoplasm is genetically characterized by 11q13 translocations and rearrangement of the BCL1 region leading to a constitutive overexpression of cyclin D1, which plays an important pathogenetic role in the development of the tumor. MCL has been considered a very aggressive disease (1). However, recent studies have identified patients with a more indolent evolution (2-8). The pathological substrate of these indolent cases is diverse. A subgroup of these patients have a distinct subtype of MCL named leukemic non-nodal MCL (nnMCL) characterized by leukemic expression, frequent splenomegaly with no or minimal nodal involvement, which typically lacks the expression of sex determining region Y-box 11 (SOX11), a transcription factor highly expressed in conventional MCL (cMCL) (2,4,5,9-12). MCL is considered to be incurable with current therapies. Nevertheless, new management strategies are improving the outcome of the patients (13). A better understanding of the molecular pathogenesis and genetic basis of the disease will allow the implementation of more effective and specific therapeutic approaches, which may help to overcome the resistance of this aggressive disease.
Epidemiology
MCL represents 3% to 10% of all lymphomas (1) and occurs predominantly in elderly men (male-to-female ratio ≥2:1) with a median age of about 60 years (range, 29 to 85 years) (14). The role of genetic susceptibility in MCL is not well described (15). Epidemiological studies have reported a two-fold significant increase of hematological neoplasms among first-degree relatives of MCL patients (16). Germline mutations in ATM and CHK2 have been detected in some patients but these genes are not involved in the few studied families with lymphoid neoplasms and MCL (17).
Postulated cell of origin and MCL subtypes
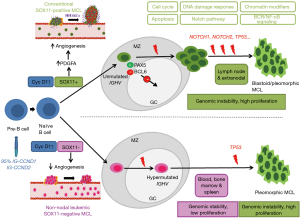
The initial oncogenic event in MCL is the t(11;14)(q13;q32) translocation that is acquired in immature pre-B cells in the bone marrow. However, the full oncogenic potential of this aberration seems to develop in mature B-cells that tend to grow in the mantle zone area of secondary lymphoid follicles. This peripheral B-cell may follow two different molecular pathways, which configure two distinctive clinical and biological subtypes of the disease (15,18) (Figure 1). The most common subtype of MCL is the cMCL, this subtype of MCL expresses the transcription factor SOX11 and originates in a B-cell that has not been exposed to the germinal center microenvironment. These tumors have no or have very low number of IGHV somatic mutations and maintain an epigenetic methylation signature reminiscent of naïve B-cells (naïve-like). This subtype develops increasing chromosomal instability with high number of chromosomal aberrations, and accumulate additional oncogenic events targeting cell cycle and DNA damage related genes. Clinically, these tumors develop generalized lymphadenopathy and usually have an aggressive clinical course. The nnMCL derives from a cell, which has experienced the germinal center and, consequently, they have high number of IGHV somatic mutations and maintain a methylation signature of memory B-cells (memory-like). These tumors also carry stable karyotypes, are SOX11-negative and clinically present with leukemic disease and frequent splenomegaly but nodal involvement is minimal or absent. The clinical course may be indolent for long periods although eventually some tumors progress with more aggressive behavior (1,7,20-22). Although the two MCL subtypes originates from mature B-cells at different stages of differentiation, the bias in IG gene usage and the presence of IG-stereotyped sequences in 10% of the cases indicate the influence of antigen selection in the clonal expansion of tumor cells in both subtypes of MCL (7,23).
Morphology
The histological features of MCL encompass a relatively large spectrum of architectural and cytological variants that may raise the differential diagnosis with other entities and are associated with particular clinical and biological characteristics.
Architectural patterns
Lymph nodes are involved with three different patterns, mantle zone, nodular and diffuse. In the mantle zone pattern the neoplastic cells expand the follicle mantle areas and surround a reactive “naked” germinal center (24,25). In these cases, the nodal architecture can be preserved. Therefore, the differential diagnosis with follicular or mantle cell hyperplasias can be challenging. However, cyclin D1 immunostaining helps to identify the cyclin D1-positive tumor cells expanding the mantle areas. Nodular MCL can represent an expansion by tumor cells of the primary follicles or a colonization and obliteration of the reactive germinal center by the tumor cells. When the nodular pattern is very prominent a morphologic differential diagnosis with a follicular lymphoma (FL) should to be considered (26). In these cases, the immunophenotypic studies help to establish the correct diagnosis. In the diffuse pattern, residual germinal centers are identified only focally. Transitional areas between nodular and diffuse patterns are common.
Cytological variants
Classic or typical MCL is characterized by a monomorphic lymphoid proliferation composed of small to medium size lymphoid cells with irregular nuclear contours with condensed chromatin, inconspicuous nucleoli and scant cytoplasm (1) (Figure 2A). Large neoplastic cells with prominent nucleoli and abundant cytoplasm are absent or very rare. The presence of these cells may correspond to residual germinal center cells or, if more abundant, should raise the differential diagnosis of other lymphoid neoplasms such as chronic lymphocytic leukemia (CLL) or FL. MCL typically show scattered epithelioid histiocytes with eosinophilic cytoplasm, which do not contain phagocytosed apoptotic bodies. Well-formed microgranulomas are not usually observed. Small vessels with fine hyalinized walls may be seen dispersed in the tumor.
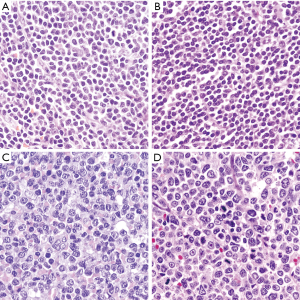
The small cell variant is composed of small and round lymphocytes with slight nuclear irregularity (Figure 2B). This variant may suggest the diagnosis of CLL but the absence of prolymphocytes, paraimmunoblasts, and proliferation growth centers help in the differential diagnosis. Proliferative activity in classic and small cell MCL is variable but usually low (<1 to 2 mitoses per high-power field). However, some cases with classic morphology can show a high mitotic index, which is associated with a worse prognosis (27,28).
More aggressive MCL include two cytological variants, blastoid and pleomorphic (29). The tumor cells in the blastoid MCL have a medium size, with rounded nuclei, finely dispersed chromatin, inconspicuous nucleoli and scant cytoplasm resembling lymphoblastic lymphoma or acute myeloid leukemias (29,30) (Figure 2C). Pleomorphic MCL is composed of a heterogeneous population of large atypical cells with ovoid or irregular, cleaved nuclei, finely dispersed chromatin and small, distinct nucleoli (31,32) (Figure 2D). MCL with pleomorphic morphology may be misdiagnosed as diffuse large B cell lymphoma (DLBCL). However, in contrast to DLBCL, pleomorphic MCL cells may have irregular nuclear contours with finely dispersed chromatin, and certain discordance between the large nuclei and relatively small nucleoli is characteristic. Ancillary studies are needed to establish the correct diagnosis. Pleomorphic and blastoid variants share the high proliferation index and the aggressive clinical course (33). WHO classification recommends distinguishing both variants based only on the morphologic features, while identification of high proliferation is not enough to classify a case as blastoid or pleomorphic since some cases of classical MCL can have high proliferation index by Ki67 (34,35). When both classic and blastoid features are identified, it is recommended reporting both cytological variants and classifying these lymphomas as blastoid or pleomorphic subtype (29). The growth pattern of blastoid and pleomorphic variants is typically diffuse, more rarely nodular or mantle zone pattern. An “in situ” pattern of blastoid or pleomorphic MCL has not yet been described (36).
In some MCL the tumor cells may have an abundant pale cytoplasm mimicking marginal zone lymphomas or monocytoid cells. This marginal zone-like variant may be associated with classic or blastoid nuclear morphology (37). Ancillary tests are mandatory in these cases including cyclin D1 and CD5 immunohistochemical evaluation. Some MCL may be associated with clusters of plasma cells. In most instances these cells are reactive with a polytypic expression of immunoglobulin light chains. However, some rare cases of MCL may show clonal plasma cell differentiation. In these cases, the plasma cell component may be clonally related or unrelated to the MCL cells (38). In cases with clonally related mature plasma cells or cells with lymphoplasmacytic differentiation these cells are cyclin D1-positive but are usually SOX11-negative and often present an indolent clinical behavior (8,38,39) (Figure 3).
MCL involvement in different organs
MCL frequently presents as a disseminated disease at diagnosis. Although the lymph nodes are the most commonly involved site, peripheral blood, bone marrow, spleen, gastrointestinal tract and other extranodal sites are also frequent sites involved by the disease.
Spleen
Splenic involvement by MCL is common and usually shows a generalized micronodular macroscopically pattern. Histologically, the tumor cells expand the white pulp but also infiltration of the red pulp can be observed (40). The nodules may be large and confluent. Residual “naked” germinal centers can be seen in around 50% of cases. The cytological appearance is similar to that seen in other locations. In some cases, a marginal zone-like area at the periphery of the nodules is observed, composed of tumor cells with abundant pale cytoplasm (40). These cases should be differentiated from marginal zone lymphoma.
Bone marrow and peripheral blood
Bone marrow infiltration is frequently seen in MCL (50% to 90% of cases) (41-43). In bone marrow biopsies, MCL can show a nodular, interstitial, paratrabecular or diffuse infiltration patterns, or a combination of them (44). The degree or histological pattern of bone marrow infiltration does not correlate with the cytological variant, nodal architectural pattern or outcome (44). Peripheral blood involvement in MCL is very common. Flow cytometry may detect leukemic MCL cells in 90% of the cases although in 15% of them the tumor cells may be scant and not observed in routine morphological examination (45). Tumor cells in peripheral blood show a cytological appearance similar to the spectrum observed in tissue samples. Most cases may have a combination of small to medium-sized cells with scant cytoplasm, nuclear irregularities, and reticular chromatin. Some cases may have cells with rounded nuclear contours. However, these cases do not show the clumped chromatin typically seen in CLL. Leukemic blastoid MCL shows medium to large cells with high nuclear-to-cytoplasmic ratio, fine dispersed chromatin and small or inconspicuous nucleoli resembling acute leukemia. Some of these cases may have MYC rearrangements (46,47). Leukemic pleomorphic MCL have very large atypical cells with prominent nucleoli. These cases may be confused with B-prolymphocytic leukemia. Cytogenetic and/or molecular studies to rule out the presence of the t(11;14)(q13;q32) or cyclin D1 overexpression are required to properly diagnose these cases as leukemic MCL (48-50).
Gastrointestinal tract
Extranodal involvement is frequent in MCL, mainly involving the gastrointestinal tract (10% to 25% of patients). Some patients can present as multiple polyps in small and large bowel (lymphomatoid polyposis) (51), but this clinical presentation is not specific of MCL (52,53). Subclinical microscopic gastrointestinal involvement by MCL cells is also common, but usually it does not have clinical implications (54). In some cases, tumor cells can infiltrate the glands mimicking lymphoepithelial lesions, making the distinction between MCL and marginal zone lymphomas difficult. Gastric involvement by nnMCL associated with Helicobacter pylori infection may regress after antibiotic treatment (5).
Immunophenotype
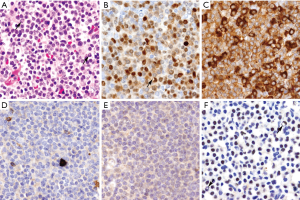
MCL expresses a mature B-cell phenotype with intense CD19, CD20, CD22, PAX5, and CD79a and the surface immunoglobulins IgM and IgD, usually with lambda light chain restriction (Figure 4A) (55). Characteristically the tumor cells co-express CD5, although it may be negative up to 10% of cMCL and 25–50% in the nnMCL subtype (Figure 4B) (7,8,15). The tumor is also positive for BCL2, CD43 and FMC7 (56), while most cases are negative for CD23, CD10, BCL6, CD200 and LEF1. Expression of the germinal center markers CD10 and BCL6 may occasionally be detected in blastoid or pleomorphic morphological variants (57-60). CD200 expression is negative in most cases, particularly cMCL but can be seen more commonly in nnMCL cases (5,61).
Cyclin D1
Expression of cyclin D1 is a constitutive and highly specific phenomenon in MCL very useful for its diagnosis. This expression is due to its rearrangement with IG genes, particularly IGHV present in around 95% of the cases (31,42,62) (Figure 4C). Cyclin D1 may be undetectable by immunohistochemistry in rare MCL carrying the t(11;14) translocation due to mutations of the C-terminus or exclusively expressing an isoform that lacks exon 5 (cyclin D1 isoform b) that render the protein undetectable by the antibody (63). Cyclin D1 expression in B-cell neoplasms is not exclusive of MCL. It can be expressed in around 25% of multiple myelomas with the t(11;14)(q13;q32), CCND1 gene amplifications, or without apparent structural alterations of the gene (64,65). Low levels of cyclin D1 are also detected in hairy-cell leukemia (66,67), and in cells of the proliferation centers in CLL, but not associated with the t(11;14)(q13;q32). Cyclin D1 expression has also been identified in around 1% of DLBCL. However, these DLBCL do not carry the t(11;14)(q13;q32) and are SOX11-negative (68). Cyclin D1 expression has been recently observed in three primary mediastinal large B cell lymphoma associated with gains of the genes but without rearrangements. These cases were SOX11-negative. Intriguingly, two of these tumors did not have apparent mediastinal involvement although they had the gene expression signature of this subtype of large B cell lymphomas (69). Cyclin D1 expression may also be detected in some peripheral T cell lymphomas (PTCL), particularly in 24% of ALK-positive anaplastic large cell lymphoma (ALCL), 7% ALK-negative ALCL, and 4% of PTCL, no otherwise specified. Cyclin D1 is negative in angioimmunoblastic T-cell lymphoma, T/NK and cutaneous T-cell lymphomas (70).
SOX11
SOX11 is a neural transcription factor highly expressed in most MCL and it is virtually negative in most mature lymphoid neoplasms (9-11). SOX11 was also identified as one of the most representative genes differentially expressed in cases with aggressive behavior but not or very lowly expressed in a subset of patients with indolent course (2). The expression of SOX11 in this tumor was surprising since it was not detected in normal lymphoid cells and in virtually any other mature lymphoid neoplasm (Figure 4D). SOX11 expression is also seen in “in situ” mantle cell neoplasias suggesting that upregulation of this transcription factor is an early event in MCL (71,72). SOX11 is also expressed in the uncommon MCL that are cyclin D1 negative (9,10,71,73). The detection of SOX11 in these cases has facilitated its recognition and better characterization (see below). Recent studies have revealed the oncogenic role of SOX11 in MCL through different pathways that include interference with the differentiation of the B-cells, increase BCR signaling and by facilitating diverse interactions of the tumor cells with the microenvironment that promote their aggressiveness (74).
In addition to MCL, SOX11 is expressed in around 30% of Burkitt lymphomas and in a high proportion of B and T-lymphoblastic lymphomas and some T-prolymphocytic leukemias, although the number of cases studied is still limited (9,10). Interestingly, SOX11 has been detected in 50% of hairy-cell leukemias that also express cyclin D1, although do not carry the t(11;14) translocation (75). On the contrary, SOX11 is not expressed in plasma cell myeloma expressing cyclin D1 and carrying the t(11;14) translocation (9,75).
Other markers
Expression of the plasma cell associated transcription factors BLIMP1 and XBP1 may be seen in around 30% of the cases, some of them with plasmacytic differentiation. These cases are usually SOX11-negative (8) (Figure 3). MYC is detected in most MCL in a low number of cells but a subset of cases, particularly with blastoid morphology and high Ki67 index, may have high expression similar to Burkitt lymphoma (76,77). This high expression may be associated with gene translocations, but not in all cases (76-78). High levels of MYC expression at protein and mRNA levels are associated with poor outcome (76,77). TP53 expression in high number of cells may be detected in around 30% of the cases. This expression is more common in blastoid and pleomorphic variants and it is associated with gene mutations and poor outcome of the patients (29,79,80).
Genetic alterations
Translocation t(11;14)(q13;q32)
The characteristic cytogenetic alteration in MCL is the t(11;14)(q13;q32), which juxtaposes the immunoglobulin heavy-chain joining region in chromosome 14 to a region on 11q13 upstream of CCND1. This translocation is present in ≥95% of cases and it is thought to represent the primary genetic event in MCL (81-85). Variant translocations involving CCND1 and the immunoglobulin light chain genes have also been reported but they are very unusual (46). These cases can be initially detected using break-apart probes for CCND1 followed by the specific fusion probes for kappa or lambda (73). In addition to MCL, the t(11;14)(q13;q32) can be found in 5% of multiple myelomas also associated with cyclin D1 overexpression (86,87). CCND1 amplifications without translocation have been observed in cases of multiple myeloma but not in MCL (64).
Secondary genetic alterations
More than 90% of MCLs display highly altered genomes, with gains/amplifications and homozygous/heterozygous losses, and other non-recurrent chromosomal rearrangements. These alterations are more common in SOX11-positive cMCL than in the nnMCL. Several chromosomal and DNA array based techniques in MCL have revealed frequent losses (1p, 2q, 6q, 8p, 9p, 9q, 10p, 11q, 13q, 17p, and 19p) and gains (3q, 7p, 8q, 10p, 11q, 12q, 13q, 15q, and 18q) (19,46) (Table 1). These chromosomal alterations target different genes relevant in MCL pathogenesis (review in another manuscript of this issue). In brief, most of the involved genes are related to cell cycle regulation (e.g., RB1 at 13q, CDKN2A at 9p21, BMI1 at 10p12, CDK4 at 12q14), DNA damage response pathway (e.g., ATM at 11q22, MDM2 at 12q15, TP53 at 17p13), and promoting cell survival (e.g., BCL2 at 18q21, TNFAIP3 at 6q23, and BIRC3 at 11q22) (46).
Table 1
| Chromosome region* | Frequency (%) | Suggested target genes | Functional process |
|---|---|---|---|
| Gains | |||
| 3q26.1-q26.32 | 28–50 | ||
| 7p22.1-p22.3 | 8–31 | IGF2BP3 (IMP3) | Insulin-like growth pathway |
| 8q24.21 | 6–32 | MYC | Cell growth, proliferation, apoptosis |
| 10p12.2-p12.31 | 6–12 | BMI1 | Cell cycle, DNA damage response |
| 11q13.3-q21 | 4–14 | CCND1 | Cell cycle |
| 12q14-q15 | 3–7 | CDK4, MDM2 | Cell cycle, DNA damage response, apoptosis |
| 13q31.3 | 5–11 | MIR17HG | Cell cycle, apoptosis |
| 15q23 | 10–23 | ||
| 18q21.33 | 3–17 | BCL2 | Apoptosis |
| Losses | |||
| 1p32.3-p33 | 18–52 | CDKN2C, FAF1 | Cell cycle, apoptosis |
| 2q13 | 3–17 | BCL2L11‡ | Pro-apoptosis |
| 2q37.1 | 15–33 | SP100-SP140 | DNA damage response |
| 6q23.3-q25 | 19–36 | TNFAIP3/LATS1 | NF-kB inhibitor/cell cycle |
| 8p21-pter | 17–34 | MCPH1/FBXO25 | DNA damage/apoptosis |
| 9p21 | 10–36 | CDKN2A, CDKN2B, MTAP/MOBKL2B | Cell cycle |
| 9q22.2-q22.31 | 17–31 | CDC14B, FANCC, GAS1 | |
| 10p14-p13 | 18–28 | ||
| 11q22.3 | 11–57 | ATM BIRC3 | DNA damage response |
| 13q13.3 | 25–55 | DLEU1, DLEU2, RB1 | Cell cycle, apoptosis |
| 13q34 | 16–54 | CUL4A, ING1, | Cell cycle, DNA damage response |
| 17p13 | 21–45 | TP53 | Cell cycle, DNA damage response |
| 19p13.3 | 10–24 | MOBKL2A | Hippo signaling pathway |
| Somatic mutations | |||
| 11q22.3 | 41–61 | ATM | DNA repair/genomic integrity |
| 17p13.1 | 14–31 | TP53 | Cell cycle, DNA damage |
| 11q13.3 | 14–34 | CCND1 | Cell cycle |
| 12q13.12 | 12–23 | KMT2D (MLL2) | Epigenetic modifier |
| 7q36.1 | 5–16 | KMT2C (MLL3) | Epigenetic modifier |
| 4p16.3 | 10–13 | NSD2 (WHSC1, MMSET) | Epigenetic modifier |
| 11q22.2 | 6–10 | BIRC3 | NF-kB signaling pathway |
| 6p21.1 | 5 | NFKBIE | NF-kB signaling pathway |
| 9q34.3 | 7 | TRAF2 | NF-kB signaling pathway |
| 9q34.3 | 5–14 | NOTCH1 | Notch signaling pathway |
| 1p12 | 5 | NOTCH2 | Notch signaling pathway |
| 8q22.3 | 7–18 | UBR5 | Ubiquitin-proteasome system |
| 7p22.2 | 3–15 | CARD11 | B-cell receptor signaling pathway |
| 1p21.2 | 3–15 | S1PR1 | Lymphocyte migration |
| 5q14.3 | 3–7 | MEF2B | Transcription factor |
| 4q31.3 | 7 | TLR2 | Toll-like receptor |
*, minimal altered regions vary slightly among different studies; ‡, homozygous deletions have been identified.
Mutational profile
Next-generation sequencing studies, including whole-exome sequencing (WES), whole-genome sequencing (WGS), RNA sequencing, and targeted sequencing have recently revealed a complex mutational landscape in MCL. Apart from the tumor suppressor genes frequently deleted and mutated in MCL such as ATM (41–61%) and TP53 (14–31%), other genes have been identified (88,89). The most commonly altered genes are activating mutations of NOTCH1 and NOTCH2, KTM2D (MLL2), KTM2C (MLL3), NSD2 (WHSC1) and MEF2B. More rarely, mutations in genes in the NF-kB signaling pathway such as CARD11, BIRC3, NFKBIE, TRAF3 as well as UBR5, and S1PR1 genes have been described (88-92).
In situ mantle cell neoplasm
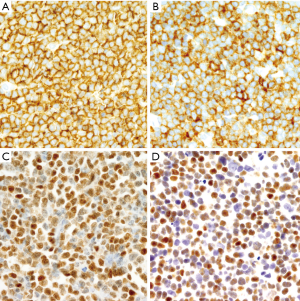
The development of MCL may follow different steps that are important to recognize because the management of the patients must be adjusted according to the phase of the disease (71,93). Cells carrying the t(11;14)(q13;q32) have been detected at very low levels with sensitive techniques in the blood of healthy individuals (8%) (94). These clones can persist for a long time, but they show an extremely low potential to convert into an overt lymphoma (95). Cyclin D1-positive cells carrying the t(11;14)(q13;q32) have been incidentally found in the mantle zones of otherwise reactive lymphoid tissues in healthy individuals (71). This pathological finding was initially named “in situ MCL” but their malignant potential seems very limited and the alternative term of “in situ mantle cell neoplasia” (isMCN) has been proposed to avoid unnecessary treatment (1). The cells are seen principally within the inner layers of the mantle cuffs of normal appearing follicles, usually intermingled with negative lymphocytes and there is no expansion of the mantle zone (71,72) (Figure 5A). The frequency of the progression from isMCN to overt MCL seems low (1/12 cases) (71) with a long latency period (72) and do not require therapy. In addition, some patients with nodal isMCN may have clonal lymphocytosis with cyclin D1 positive cells (71).
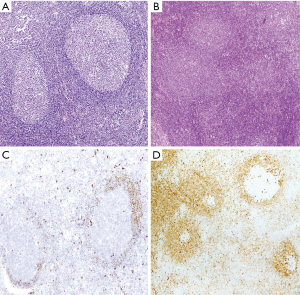
Most in situ lesions are SOX11-positive, whereas few are SOX11-negative. These findings suggest that the in situ stage may be a common step in both SOX11-negative and SOX11-positive subtypes of MCL (71,96). It is important to differentiate isMCN from early involvement by overt MCL with a mantle zone growth pattern (Figure 5B), as this situation correlates with progression to disseminated disease more frequently than in situ lesions. In these cases, the mantles are usually expanded and densely occupied by cyclin D1-positive cells that may focally extend to interfollicular areas (71,72) (Figure 5C,D).
Tumor progression
In general, the histological pattern of MCL remains stable in sequential biopsies (30,97,98). In some cases, a progression from a nodular pattern to a diffuse pattern is observed (97,98), while in others the histologic patterns change in the course of the disease in successive biopsies (99). Interestingly, 22% of MCL cases with classic/small cell morphology at diagnosis may progress to a blastoid or pleomorphic variant in the subsequent relapse (97,99,100). However, half of MCL with blastoid morphology can recur as a classical variant (100). The proliferation index evaluated by Ki67 increases over time and it is associated with prognosis in the primary and the relapse biopsy specimens (100). Overt leukemic involvement can present at diagnosis while in other patients, peripheral blood involvement may appear during the course of the disease representing tumor progression.
Cyclin D1-negative MCL
Rare cases with the morphology and phenotype of MCL are negative for cyclin D1 expression and do not carry the t(11;14)(q13;q32). These cases are considered a molecular variant of MCL since they have a similar global gene expression profile and clinical characteristics (20,101). These cases also express SOX11 and the evaluation of this biomarker is very useful in the identification of these tumors in the clinical practice (73,88). Cyclin D1-negative MCL may have the same architectural and cytological variants as cyclin D1-positive tumors including cases with mantle zone pattern or blastoid/pleomorphic cytology (20,101).
Recent genetic and molecular studies have identified that virtually all cyclin D1-negattive MCL carry CCND2/CCND3 rearrangements with IG genes, particularly with kappa and lambda light chains (20). Intriguingly, some cases carry cryptic insertions of the kappa and lambda enhancer regions in the vicinity of CCND2 and CCND3 that are also associated with high overexpression of the genes. These cryptic translocations may be recognized in routine formalin fixed, paraffin embedded tissues using specific FISH probes but given the small size of the enhancer region the juxtaposition is better detected in captured images. Intriguingly, a small subset of cases with morphological, and phenotypic features of MCL, including SOX11 expression, do not overexpress any of these three cyclin D but have up-regulation of CCNE1 and CCNE2 without apparent genetic alterations (102). Hence, the combined study of SOX11 expression together with the study of CCND2/CCND3 rearrangements by FISH, particularly with break-apart probes and specific probes for the light chain enhancer regions, and also the quantification of CCND2/D3/E1/E2 mRNA levels by qPCR may be useful tools for the identification of these cases (103).
Leukemic NNMCL
The nnMCL has been recently accepted as a subtype of MCL in the updated 2016 WHO classification of lymphoid neoplasms (1). These tumors usually have an initial indolent behavior with a clinical presentation as leukemic disease with no or minimal lymphadenopathy and frequent splenomegaly (1-8). These cases frequently show small cell morphology, resembling CLL, are SOX11-negative, may express CD200 and CD5 may be negative in 25–50% of the cases (1,5,7). Contrary to cMCL, most of these tumors have mutated IGHV and have simple karyotypes with no o very few chromosomal alterations in addition to the t(11;14)(q13;q32) (2,3). Despite these clinical and biological differences, the global genome expression profile of these indolent nnMCL is more similar to cMCL than to other subtypes of leukemic lymphoid neoplasms suggesting that they correspond to a molecular subtype of the disease (2). The identification of cyclin D1-positive “in situ” mantle cell lesions in which the cells are SOX11-negative also suggests that nnMCL is a subtype of MCL (71). The nnMCL and cMCL variants have also clear differences in the gene expression of certain programs. Expression profiling and experimental studies suggest that nnMCL do not have the tumor invasion properties and angiogenic potential seen in cMCL driven by SOX11 expression (104,105).
Robust criteria to distinguish these MCL subtypes in the clinical practice and additional biological parameters that influence their evolution are not well defined. SOX11 is currently used in the proper clinical context to identify both subtypes. However, SOX11 expression is usually studied by immunohistochemistry in biopsies and most nnMCL are leukemic with no tissue samples available. Recently a novel molecular assay based on the gene expression of 16 genes has been developed that reliably distinguishes cMCL and nnMCL using blood samples. This study confirmed that cases assigned to the nnMCL and cMCL differed in nodal presentation, lactate dehydrogenase, immunoglobulin heavy chain gene mutational status, management options, genomic complexity, and CDKN2A/ATM deletions, but the proportion with 17p/TP53 aberrations was similar in both subgroups. nnMCL had a significant better overall survival from the time of diagnosis than cMCL (3-year OS 92% vs. 69%; P=0.006) and longer time to first treatment. In addition, genomic complexity and TP53/CDKN2A aberrations were associated with shorter OS in the entire series and both cMCL and nnMCL subtypes. Therefore, the combination of this assay with genetic alterations may recognize the two subtypes of MCL and may provide useful biological information for the management of the patients (21,106).
Prognostic parameters
Several clinical, pathological and biological parameters have been investigated to evaluate the possibility of predicting the heterogeneous evolution of patients with MCL. The most consolidated clinical prognostic model is the MCL International Prognostic Index (MIPI) that combines age, performance status, LDH, and lymphocyte counts (107). This model is improved by incorporating the proliferative index measured by the Ki67 antigen (biological MIPI) (108). A high proliferative index evaluated by Ki67 immunohistochemical staining has been associated with a worse outcome, even in patients treated in randomized trials with immunochemotherapy and high dose regimens (35,107). Morphological parameters such as the architectural nodular and diffuse pattern and the cytological blastoid/pleomorphic variants have classically been associated with prognosis. However, their value is not independent of the Ki67 index (107). This is in part because of some MCL with classic morphology that have high proliferation show poor outcome (36).
In spite of the relevant clinical value, the evaluation of the Ki67 immunohistochemistry has some limitations in terms of intra and interobserver reproducibility. Recommendations and guidelines to assess the proliferation activity by Ki67 include the evaluation only in nodal tissue samples, count at least five high power fields, and avoid counting in residual reactive germinal centers, hot spots of proliferation, and accompanying T-cells (35).
The relevance of the proliferative activity in predicting outcome in MCL was also highlighted in the early gene expression profiling studies using RNA extracted from frozen tissues. These studies identified an expression signature composed of genes related to proliferation that discriminated different risk groups of patients with MCL (109). Recently, this proliferation signature has been adapted in a new assay useful for RNA extracted from formalin fixed and paraffin embedded tissues (110). The assay uses the NanoString platform and is composed of 35 genes, 17 associated with proliferation and 18 housekeeping genes (MCL35 assay) and its results are highly reproducible among laboratories (110). The assay assigns patients to high, standard, and low-risk groups, with median overall survival of 1.1, 2.6, and 8.6 years, respectively. The MCL35 proliferation signature score significantly correlates with the Ki67 index and improves its prognostic prediction. These results have been confirmed in MCL patients treated in different clinical trials (111,112).
Early genetic and comparative genomic hybridizations studies recognized the prognostic value of complex karyotypes and high genomic complexity associated with a more aggressive clinical behavior (113-115). Several individual chromosomal alterations including 3q gains, and deletions of 9p and 17p targeting CDKN2A and TP53 respectively, correlate also with poor outcome (115). The prognostic value of TP53 and CDKN2A alterations has been recently confirmed in several clinical trials (28). TP53 mutations and protein overexpression (>50% of cells) are associated with a very aggressive behavior that is not overcome by treatments using intensive regimens (29,116). Similar to TP53 aberrations CDKN2A deletions confer a dismal prognosis with a median OS lower than 2 years and seems to add independent value to the TP53 aberrations (117).
Conclusions
The spectrum of pathological and clinical characteristics of MCL has expanded in recent years. The diagnosis of this entity requires the integration of morphologic and immunophenotypic studies and sometimes the use of genetic and molecular studies may be required to clarify the differential diagnosis and predict the evolution of the disease. The diverse evolution of the patients that may vary from indolent to very aggressive and the development of new therapeutic strategies require a precise stratification of the patients in different risk groups to personalize the best management possible. The better understanding of the molecular basis of the disease together with new technologies that allow transferring this knowledge into the clinics and the development of novel therapies are opening new perspectives that should result in an improved outcome of the patients.
Acknowledgments
Funding: Dr. Luis Veloza was supported by Colciencias (Colombian Department of Science, Technology and Innovation) from Government of Colombia through the international doctoral fellowship (No. 728-2015). Dr. Elias Campo research is supported by the Ministerio de Economía y Competitividad (grant No. SAF2015-64885-R).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Martin Dreyling) for the series “Future Directions for Mantle Cell Lymphoma” published in Annals of Lymphoma. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aol.2019.03.01). The series “Future Directions for Mantle Cell Lymphoma” was commissioned by the editorial office without any funding or sponsorship. Luis Veloza reports personal fees from Colombian Department of Science, Technology and Innovation, during the conduct of the study. Elias Campo reports grants from NIH, grants from Spanish Ministry of Science, grants from Generalitat de Catalunya, grants from Spanish Ministry of Health, grants and personal fees from Generalitat de Catalunya, during the conduct of the study; personal fees from Takeda, personal fees from Ilumina, personal fees from NanoString, personal fees from Janssen, personal fees from Roche, grants and personal fees from Gilead, grants from European Research Council, outside the submitted work; In addition, Elisa Campo has a patent NanoString Technologies licensed. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016;127:2375-90. [Crossref] [PubMed]
- Fernàndez V, Salamero O, Espinet B, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res 2010;70:1408-18. [Crossref] [PubMed]
- Royo C, Navarro A, Clot G, et al. Non-nodal type of mantle cell lymphoma is a specific biological and clinical subgroup of the disease. Leukemia 2012;26:1895-8. [Crossref] [PubMed]
- Ondrejka SL, Lai R, Smith SD, et al. Indolent mantle cell leukemia: a clinicopathological variant characterized by isolated lymphocytosis, interstitial bone marrow involvement, kappa light chain restriction, and good prognosis. Haematologica 2011;96:1121-7. [Crossref] [PubMed]
- Espinet B, Ferrer A, Bellosillo B, et al. Distinction between asymptomatic monoclonal B-cell lymphocytosis with cyclin D1 overexpression and mantle cell lymphoma: from molecular profiling to flow cytometry. Clin Cancer Res 2014;20:1007-19. [Crossref] [PubMed]
- Vegliante MC, Palomero J, Perez-Galan P, et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood 2013;121:2175-85. [Crossref] [PubMed]
- Navarro A, Clot G, Royo C, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res 2012;72:5307-16. [Crossref] [PubMed]
- Ribera-Cortada I, Martinez D, Amador V, et al. Plasma cell and terminal B-cell differentiation in mantle cell lymphoma mainly occur in the SOX11-negative subtype. Mod Pathol 2015;28:1435-47. [Crossref] [PubMed]
- Dictor M, Ek S, Sundberg M, et al. Strong lymphoid nuclear expression of SOX11 transcription factor defines lymphoblastic neoplasms, mantle cell lymphoma and Burkitt's lymphoma. Haematologica 2009;94:1563-8. [Crossref] [PubMed]
- Mozos A, Royo C, Hartmann E, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica 2009;94:1555-62. [Crossref] [PubMed]
- Ek S, Dictor M, Jerkeman M, et al. Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood 2008;111:800-5. [Crossref] [PubMed]
- Xu W, Li JY. SOX11 expression in mantle cell lymphoma. Leuk Lymphoma 2010;51:1962-7. [Crossref] [PubMed]
- Schieber M, Gordon LI, Karmali R. Current overview and treatment of mantle cell lymphoma. F1000Res 2018; [Crossref] [PubMed]
- A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin's lymphoma. The Non-Hodgkin's Lymphoma Classification Project. Blood 1997;89:3909-18. [PubMed]
- Puente XS, Jares P, Campo E. Chronic lymphocytic leukemia and mantle cell lymphoma: crossroads of genetic and microenvironment interactions. Blood 2018;131:2283-96. [Crossref] [PubMed]
- Smedby KE, Sampson JN, Turner JJ, et al. Medical history, lifestyle, family history, and occupational risk factors for mantle cell lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr 2014;2014:76-86. [Crossref] [PubMed]
- Tort F, Camacho E, Bosch F, et al. Familial lymphoid neoplasms in patients with mantle cell lymphoma. Haematologica 2004;89:314-9. [PubMed]
- Jares P, Colomer D, Campo E. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest 2012;122:3416-23. [Crossref] [PubMed]
- Beà S, Amador V. Role of SOX11 and Genetic Events Cooperating with Cyclin D1 in Mantle Cell Lymphoma. Curr Oncol Rep 2017;19:43. [Crossref] [PubMed]
- Salaverria I, Royo C, Carvajal-Cuenca A, et al. CCND2 rearrangements are the most frequent genetic events in cyclin D1(-) mantle cell lymphoma. Blood 2013;121:1394-402. [Crossref] [PubMed]
- Clot G, Jares P, Gine E, et al. A gene signature that distinguishes conventional and leukemic nonnodal mantle cell lymphoma helps predict outcome. Blood 2018;132:413-22. [Crossref] [PubMed]
- Queirós AC, Beekman R, Vilarrasa-Blasi R, et al. Decoding the DNA Methylome of Mantle Cell Lymphoma in the Light of the Entire B Cell Lineage. Cancer Cell 2016;30:806-21. [Crossref] [PubMed]
- Hadzidimitriou A, Agathangelidis A, Darzentas N, et al. Is there a role for antigen selection in mantle cell lymphoma? Immunogenetic support from a series of 807 cases. Blood 2011;118:3088-95. [Crossref] [PubMed]
- Weisenburger DD, Kim H, Rappaport H. Mantle-zone lymphoma: a follicular variant of intermediate lymphocytic lymphoma. Cancer 1982;49:1429-38. [Crossref] [PubMed]
- Majlis A, Pugh WC, Rodriguez MA, et al. Mantle cell lymphoma: correlation of clinical outcome and biologic features with three histologic variants. J Clin Oncol 1997;15:1664-71. [Crossref] [PubMed]
- Vadlamudi G, Lionetti KA, Greenberg S, et al. Leukemic phase of mantle cell lymphoma two case reports and review of the literature. Arch Pathol Lab Med 1996;120:35-40. [PubMed]
- Dreyling MH, Bullinger L, Ott G, et al. Alterations of the cyclin D1/p16-pRB pathway in mantle cell lymphoma. Cancer Res 1997;57:4608-14. [PubMed]
- Pinyol M, Cobo F, Bea S, et al. p16(INK4a) gene inactivation by deletions, mutations, and hypermethylation is associated with transformed and aggressive variants of non-Hodgkin's lymphomas. Blood 1998;91:2977-84. [PubMed]
- Aukema SM, Hoster E, Rosenwald A, et al. Expression of TP53 is associated with the outcome of MCL independent of MIPI and Ki-67 in trials of the European MCL Network. Blood 2018;131:417-20. [Crossref] [PubMed]
- Lardelli P, Bookman MA, Sundeen J, et al. Lymphocytic lymphoma of intermediate differentiation. Morphologic and immunophenotypic spectrum and clinical correlations. Am J Surg Pathol 1990;14:752-63. [Crossref] [PubMed]
- Bosch F, Jares P, Campo E, et al. PRAD-1/cyclin D1 gene overexpression in chronic lymphoproliferative disorders: a highly specific marker of mantle cell lymphoma. Blood 1994;84:2726-32. [PubMed]
- Ott MM, Ott G, Kuse R, et al. The anaplastic variant of centrocytic lymphoma is marked by frequent rearrangements of the bcl-1 gene and high proliferation indices. Histopathology 1994;24:329-34. [Crossref] [PubMed]
- Shrestha R, Bhatt VR, Guru Murthy GS, et al. Clinicopathologic features and management of blastoid variant of mantle cell lymphoma. Leuk Lymphoma 2015;56:2759-67. [Crossref] [PubMed]
- Determann O, Hoster E, Ott G, et al. Ki-67 predicts outcome in advanced-stage mantle cell lymphoma patients treated with anti-CD20 immunochemotherapy: results from randomized trials of the European MCL Network and the German Low Grade Lymphoma Study Group. Blood 2008;111:2385-7. [Crossref] [PubMed]
- Klapper W, Hoster E, Determann O, et al. Ki-67 as a prognostic marker in mantle cell lymphoma-consensus guidelines of the pathology panel of the European MCL Network. J Hematop 2009;2:103-11. [Crossref] [PubMed]
- Dreyling M, Klapper W, Rule S. Blastoid and pleomorphic mantle cell lymphoma: still a diagnostic and therapeutic challenge! Blood 2018;132:2722-9. [Crossref] [PubMed]
- Swerdlow SH, Zukerberg LR, Yang WI, et al. The morphologic spectrum of non-Hodgkin's lymphomas with BCL1/cyclin D1 gene rearrangements. Am J Surg Pathol 1996;20:627-40. [Crossref] [PubMed]
- Visco C, Hoeller S, Malik JT, et al. Molecular characteristics of mantle cell lymphoma presenting with clonal plasma cell component. Am J Surg Pathol 2011;35:177-89. [Crossref] [PubMed]
- Swerdlow SH, Kuzu I, Dogan A, et al. The many faces of small B cell lymphomas with plasmacytic differentiation and the contribution of MYD88 testing. Virchows Arch 2016;468:259-75. [Crossref] [PubMed]
- Piris MA, Mollejo M, Campo E, et al. A marginal zone pattern may be found in different varieties of non-Hodgkin's lymphoma: the morphology and immunohistology of splenic involvement by B-cell lymphomas simulating splenic marginal zone lymphoma. Histopathology 1998;33:230-9. [Crossref] [PubMed]
- Pittaluga S, Verhoef G, Criel A, et al. Prognostic significance of bone marrow trephine and peripheral blood smears in 55 patients with mantle cell lymphoma. Leuk Lymphoma 1996;21:115-25. [Crossref] [PubMed]
- Vasef MA, Medeiros LJ, Koo C, et al. Cyclin D1 immunohistochemical staining is useful in distinguishing mantle cell lymphoma from other low-grade B-cell neoplasms in bone marrow. Am J Clin Pathol 1997;108:302-7. [Crossref] [PubMed]
- Wasman J, Rosenthal NS, Farhi DC. Mantle cell lymphoma. Morphologic findings in bone marrow involvement. Am J Clin Pathol 1996;106:196-200. [Crossref] [PubMed]
- Cohen PL, Kurtin PJ, Donovan KA, et al. Bone marrow and peripheral blood involvement in mantle cell lymphoma. Br J Haematol 1998;101:302-10. [Crossref] [PubMed]
- Ferrer A, Salaverria I, Bosch F, et al. Leukemic involvement is a common feature in mantle cell lymphoma. Cancer 2007;109:2473-80. [Crossref] [PubMed]
- Royo C, Salaverria I, Hartmann EM, et al. The complex landscape of genetic alterations in mantle cell lymphoma. Semin Cancer Biol 2011;21:322-34. [Crossref] [PubMed]
- Hao S, Sanger W, Onciu M, et al. Mantle cell lymphoma with 8q24 chromosomal abnormalities: a report of 5 cases with blastoid features. Mod Pathol 2002;15:1266-72. [Crossref] [PubMed]
- Zoldan MC, Inghirami G, Masuda Y, et al. Large-cell variants of mantle cell lymphoma: cytologic characteristics and p53 anomalies may predict poor outcome. Br J Haematol 1996;93:475-86. [Crossref] [PubMed]
- Schlette E, Lai R, Onciu M, et al. Leukemic mantle cell lymphoma: clinical and pathologic spectrum of twenty-three cases. Mod Pathol 2001;14:1133-40. [Crossref] [PubMed]
- Dunphy CH, Perkins SL. Mantle cell leukemia, prolymphocytoid type: a rarely described form. Leuk Lymphoma 2001;41:683-7. [Crossref] [PubMed]
- O'Briain DS, Kennedy MJ, Daly PA, et al. Multiple lymphomatous polyposis of the gastrointestinal tract. A clinicopathologically distinctive form of non-Hodgkin's lymphoma of B-cell centrocytic type. Am J Surg Pathol 1989;13:691-9. [Crossref] [PubMed]
- Moynihan MJ, Bast MA, Chan WC, et al. Lymphomatous polyposis. A neoplasm of either follicular mantle or germinal center cell origin. Am J Surg Pathol 1996;20:442-52. [Crossref] [PubMed]
- Hashimoto Y, Nakamura N, Kuze T, et al. Multiple lymphomatous polyposis of the gastrointestinal tract is a heterogenous group that includes mantle cell lymphoma and follicular lymphoma: analysis of somatic mutation of immunoglobulin heavy chain gene variable region. Hum Pathol 1999;30:581-7. [Crossref] [PubMed]
- Romaguera JE, Medeiros LJ, Hagemeister FB, et al. Frequency of gastrointestinal involvement and its clinical significance in mantle cell lymphoma. Cancer 2003;97:586-91. [Crossref] [PubMed]
- Campo E, Raffeld M, Jaffe ES. Mantle-cell lymphoma. Semin Hematol 1999;36:115-27. [PubMed]
- Swerdlow SH, Utz GL, Williams ME. Bcl-2 protein in centrocytic lymphoma; a paraffin section study. Leukemia 1993;7:1456-8. [PubMed]
- Akhter A, Mahe E, Street L, et al. CD10-positive mantle cell lymphoma: biologically distinct entity or an aberrant immunophenotype? Insight, through gene expression profile in a unique case series. J Clin Pathol 2015;68:844-8. [Crossref] [PubMed]
- Camacho FI, Garcia JF, Cigudosa JC, et al. Aberrant Bcl6 protein expression in mantle cell lymphoma. Am J Surg Pathol 2004;28:1051-6. [Crossref] [PubMed]
- Gao J, Peterson L, Nelson B, et al. Immunophenotypic variations in mantle cell lymphoma. Am J Clin Pathol 2009;132:699-706. [Crossref] [PubMed]
- Morice WG, Hodnefield JM, Kurtin PJ, et al. An unusual case of leukemic mantle cell lymphoma with a blastoid component showing loss of CD5 and aberrant expression of CD10. Am J Clin Pathol 2004;122:122-7. [Crossref] [PubMed]
- Hu Z, Sun Y, Schlette EJ, et al. CD200 expression in mantle cell lymphoma identifies a unique subgroup of patients with frequent IGHV mutations, absence of SOX11 expression, and an indolent clinical course. Mod Pathol 2018;31:327-36. [Crossref] [PubMed]
- de Boer CJ, van Krieken JH, Kluin-Nelemans HC, et al. Cyclin D1 messenger RNA overexpression as a marker for mantle cell lymphoma. Oncogene 1995;10:1833-40. [PubMed]
- Iaccarino I, Afify L, Aukema SM, et al. t(11;14)-positive mantle cell lymphomas lacking cyclin D1 (CCND1) immunostaining because of a CCND1 mutation or exclusive expression of the CCND1b isoform. Haematologica 2018;103:e432-5. [Crossref] [PubMed]
- Hoechtlen-Vollmar W, Menzel G, Bartl R, et al. Amplification of cyclin D1 gene in multiple myeloma: clinical and prognostic relevance. Br J Haematol 2000;109:30-8. [Crossref] [PubMed]
- Specht K, Haralambieva E, Bink K, et al. Different mechanisms of cyclin D1 overexpression in multiple myeloma revealed by fluorescence in situ hybridization and quantitative analysis of mRNA levels. Blood 2004;104:1120-6. [Crossref] [PubMed]
- Bosch F, Campo E, Jares P, et al. Increased expression of the PRAD-1/CCND1 gene in hairy cell leukaemia. Br J Haematol 1995;91:1025-30. [Crossref] [PubMed]
- de Boer CJ, Kluin-Nelemans JC, Dreef E, et al. Involvement of the CCND1 gene in hairy cell leukemia. Ann Oncol 1996;7:251-6. [Crossref] [PubMed]
- Hsiao SC, Cortada IR, Colomo L, et al. SOX11 is useful in differentiating cyclin D1-positive diffuse large B-cell lymphoma from mantle cell lymphoma. Histopathology 2012;61:685-93. [Crossref] [PubMed]
- Chen BJ, Ruminy P, Roth CG, et al. Cyclin D1-positive Mediastinal Large B-Cell Lymphoma With Copy Number Gains of CCND1 Gene: A Study of 3 Cases With Nonmediastinal Disease. Am J Surg Pathol 2019;43:110-20. [Crossref] [PubMed]
- Song JY, Song L, Herrera AF, et al. Cyclin D1 expression in peripheral T-cell lymphomas. Mod Pathol 2016;29:1306-12. [Crossref] [PubMed]
- Carvajal-Cuenca A, Sua LF, Silva NM, et al. In situ mantle cell lymphoma: clinical implications of an incidental finding with indolent clinical behavior. Haematologica 2012;97:270-8. [Crossref] [PubMed]
- Adam P, Schiefer AI, Prill S, et al. Incidence of preclinical manifestations of mantle cell lymphoma and mantle cell lymphoma in situ in reactive lymphoid tissues. Mod Pathol 2012;25:1629-36. [Crossref] [PubMed]
- Sander B, Quintanilla-Martinez L, Ott G, et al. Mantle cell lymphoma--a spectrum from indolent to aggressive disease. Virchows Arch 2016;468:245-57. [Crossref] [PubMed]
- Beekman R, Amador V, Campo E. SOX11, a key oncogenic factor in mantle cell lymphoma. Curr Opin Hematol 2018;25:299-306. [Crossref] [PubMed]
- Chen YH, Gao J, Fan G, et al. Nuclear expression of sox11 is highly associated with mantle cell lymphoma but is independent of t(11;14)(q13;q32) in non-mantle cell B-cell neoplasms. Mod Pathol 2010;23:105-12. [Crossref] [PubMed]
- Oberley MJ, Rajguru SA, Zhang C, et al. Immunohistochemical evaluation of MYC expression in mantle cell lymphoma. Histopathology 2013;63:499-508. [PubMed]
- Hartmann E, Fernandez V, Moreno V, et al. Five-gene model to predict survival in mantle-cell lymphoma using frozen or formalin-fixed, paraffin-embedded tissue. J Clin Oncol 2008;26:4966-72. [Crossref] [PubMed]
- Hu Z, Medeiros LJ, Chen Z, et al. Mantle Cell Lymphoma With MYC Rearrangement: A Report of 17 Patients. Am J Surg Pathol 2017;41:216-24. [Crossref] [PubMed]
- Hernandez L, Fest T, Cazorla M, et al. p53 gene mutations and protein overexpression are associated with aggressive variants of mantle cell lymphomas. Blood 1996;87:3351-9. [PubMed]
- Slotta-Huspenina J, Koch I, de Leval L, et al. The impact of cyclin D1 mRNA isoforms, morphology and p53 in mantle cell lymphoma: p53 alterations and blastoid morphology are strong predictors of a high proliferation index. Haematologica 2012;97:1422-30. [Crossref] [PubMed]
- Li JY, Gaillard F, Moreau A, et al. Detection of translocation t(11;14)(q13;q32) in mantle cell lymphoma by fluorescence in situ hybridization. Am J Pathol 1999;154:1449-52. [Crossref] [PubMed]
- Rosenberg CL, Wong E, Petty EM, et al. PRAD1, a candidate BCL1 oncogene: mapping and expression in centrocytic lymphoma. Proc Natl Acad Sci U S A 1991;88:9638-42. [Crossref] [PubMed]
- Vaandrager JW, Schuuring E, Zwikstra E, et al. Direct visualization of dispersed 11q13 chromosomal translocations in mantle cell lymphoma by multicolor DNA fiber fluorescence in situ hybridization. Blood 1996;88:1177-82. [PubMed]
- Vandenberghe E, De Wolf-Peeters C, van den Oord J, et al. Translocation (11;14): a cytogenetic anomaly associated with B-cell lymphomas of non-follicle centre cell lineage. J Pathol 1991;163:13-8. [Crossref] [PubMed]
- Williams ME, Swerdlow SH, Rosenberg CL, et al. Chromosome 11 translocation breakpoints at the PRAD1/cyclin D1 gene locus in centrocytic lymphoma. Leukemia 1993;7:241-5. [PubMed]
- Raffeld M, Jaffe ES. bcl-1, t(11;14), and mantle cell-derived lymphomas. Blood 1991;78:259-63. [PubMed]
- de Boer CJ, van Krieken JH, Schuuring E, et al. Bcl-1/cyclin D1 in malignant lymphoma. Ann Oncol 1997;8:109-17. [Crossref] [PubMed]
- Beà S, Valdes-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A 2013;110:18250-5. [Crossref] [PubMed]
- Zhang J, Jima D, Moffitt AB, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood 2014;123:2988-96. [Crossref] [PubMed]
- Wu C, de Miranda NF, Chen L, et al. Genetic heterogeneity in primary and relapsed mantle cell lymphomas: Impact of recurrent CARD11 mutations. Oncotarget 2016;7:38180-90. [PubMed]
- Kridel R, Meissner B, Rogic S, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood 2012;119:1963-71. [Crossref] [PubMed]
- Meissner B, Kridel R, Lim RS, et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood 2013;121:3161-4. [Crossref] [PubMed]
- Karube K, Scarfo L, Campo E, et al. Monoclonal B cell lymphocytosis and "in situ" lymphoma. Semin Cancer Biol 2014;24:3-14. [Crossref] [PubMed]
- Lecluse Y, Lebailly P, Roulland S, et al. t(11;14)-positive clones can persist over a long period of time in the peripheral blood of healthy individuals. Leukemia 2009;23:1190-3. [Crossref] [PubMed]
- Christian B, Zhao W, Hamadani M, et al. Mantle cell lymphoma 12 years after allogeneic bone marrow transplantation occurring simultaneously in recipient and donor. J Clin Oncol 2010;28:e629-32. [Crossref] [PubMed]
- Fend F, Cabecadas J, Gaulard P, et al. Early lesions in lymphoid neoplasia: Conclusions based on the Workshop of the XV. Meeting of the European Association of Hematopathology and the Society of Hematopathology, in Uppsala, Sweden. J Hematop. 2012;5: [Crossref] [PubMed]
- Argatoff LH, Connors JM, Klasa RJ, et al. Mantle cell lymphoma: a clinicopathologic study of 80 cases. Blood 1997;89:2067-78. [PubMed]
- Swerdlow SH, Habeshaw JA, Murray LJ, et al. Centrocytic lymphoma: a distinct clinicopathologic and immunologic entity. A multiparameter study of 18 cases at diagnosis and relapse. Am J Pathol 1983;113:181-97. [PubMed]
- Norton AJ, Matthews J, Pappa V, et al. Mantle cell lymphoma: natural history defined in a serially biopsied population over a 20-year period. Ann Oncol 1995;6:249-56. [Crossref] [PubMed]
- Vogt N, Klapper W. Variability in morphology and cell proliferation in sequential biopsies of mantle cell lymphoma at diagnosis and relapse: clinical correlation and insights into disease progression. Histopathology 2013;62:334-42. [Crossref] [PubMed]
- Zeng W, Fu K, Quintanilla-Fend L, et al. Cyclin D1-negative blastoid mantle cell lymphoma identified by SOX11 expression. Am J Surg Pathol 2012;36:214-9. [Crossref] [PubMed]
- Martín-Garcia D, Navarro A, Valdés-Mas R, et al. CCND2 and CCND3 hijack immunoglobulin light-chain enhancers in cyclin D1- mantle cell lymphoma. Blood 2019;133:940-51. [Crossref] [PubMed]
- Quintanilla-Martinez L, Slotta-Huspenina J, Koch I, et al. Differential diagnosis of cyclin D2+ mantle cell lymphoma based on fluorescence in situ hybridization and quantitative real-time-PCR. Haematologica 2009;94:1595-8. [Crossref] [PubMed]
- Palomero J, Vegliante MC, Rodriguez ML, et al. SOX11 promotes tumor angiogenesis through transcriptional regulation of PDGFA in mantle cell lymphoma. Blood 2014;124:2235-47. [Crossref] [PubMed]
- Balsas P, Palomero J, Eguileor A, et al. SOX11 promotes tumor protective microenvironment interactions through CXCR4 and FAK regulation in mantle cell lymphoma. Blood 2017;130:501-13. [Crossref] [PubMed]
- Martin P. A tale of two mantle cell lymphomas. Blood 2018;132:347-8. [Crossref] [PubMed]
- Hoster E, Rosenwald A, Berger F, et al. Prognostic Value of Ki-67 Index, Cytology, and Growth Pattern in Mantle-Cell Lymphoma: Results From Randomized Trials of the European Mantle Cell Lymphoma Network. J Clin Oncol 2016;34:1386-94. [Crossref] [PubMed]
- Dreyling M, Ferrero S, Vogt N, et al. New paradigms in mantle cell lymphoma: is it time to risk-stratify treatment based on the proliferative signature? Clin Cancer Res 2014;20:5194-206. [Crossref] [PubMed]
- Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell 2003;3:185-97. [Crossref] [PubMed]
- Scott DW, Abrisqueta P, Wright GW, et al. New Molecular Assay for the Proliferation Signature in Mantle Cell Lymphoma Applicable to Formalin-Fixed Paraffin-Embedded Biopsies. J Clin Oncol 2017;35:1668-77. [Crossref] [PubMed]
- Holte H, Beiske K, Boyle M, et al. The MCL35 gene expression proliferation assay predicts high-risk MCL patients in a Norwegian cohort of younger patients given intensive first line therapy. Br J Haematol 2018;183:225-34. [Crossref] [PubMed]
- Rauert-Wunderlich H, Mottok A, Scott DW, et al. Validation of the MCL35 gene expression proliferation assay in randomized trials of the European Mantle Cell Lymphoma Network. Br J Haematol 2019;184:616-24. [Crossref] [PubMed]
- Beà S, Ribas M, Hernandez JM, et al. Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood 1999;93:4365-74. [PubMed]
- Cuneo A, Bigoni R, Rigolin GM, et al. Cytogenetic profile of lymphoma of follicle mantle lineage: correlation with clinicobiologic features. Blood 1999;93:1372-80. [PubMed]
- Salaverria I, Zettl A, Bea S, et al. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression-based proliferation signature. J Clin Oncol 2007;25:1216-22. [Crossref] [PubMed]
- Eskelund CW, Dahl C, Hansen JW, et al. TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood 2017;130:1903-10. [Crossref] [PubMed]
- Delfau-Larue MH, Klapper W, Berger F, et al. High-dose cytarabine does not overcome the adverse prognostic value of CDKN2A and TP53 deletions in mantle cell lymphoma. Blood 2015;126:604-11. [Crossref] [PubMed]
Cite this article as: Veloza L, Ribera-Cortada I, Campo E. Mantle cell lymphoma pathology update in the 2016 WHO classification. Ann Lymphoma 2019;3:3.


