
Biology of splenic and nodal marginal zone lymphomas
Introduction
Marginal zone lymphoma (MZL) comprises three different entities, as recognized by the World Health Organization (WHO) in the classification of lymphoid disorders: nodal MZL (NMZL), splenic MZL (SMZL) and extranodal MZL of mucosa-associated lymphoid tissue (MALT lymphoma) (1). NMZL and SMZL are infrequently occurring small B cell neoplasms; while NMZL originates in lymph nodes and subsequently infiltrates the spleen and bone marrow, SMZL originates in the spleen with frequent peripheral blood involvement. The postulated cell of origin is an antigen-experienced B cell, consistent with the homing B cells at the marginal zone (MZ). The normal MZ surrounds the secondary lymphoid follicles and is continuously exposed to extrinsic antigens that determine a rapid antibody response upon engagement of the B cell receptor (BCR).
The absence of diagnostic and prognostic biomarkers together with the relative rarity of these disorders makes their diagnosis and molecular investigation challenging. Genomic aberrations frequently result in the activation of survival pathways common to SMZL and NMZL. These include NF-κB activation as a result of mutation and/or deletions of TNFAIP3, mutations of chromatin remodeling genes, chromosomal trisomies (+3, +18), NOTCH pathway mutations and genomic aberrations affecting the KLF2 transcription factor (2-13). In addition to shared alterations, there are also specific aberrations that characterize each subtype. In SMZL, deletions of 7q account for 30% of all chromosomal aberrations, while gains of 3/3q, 9q,12q and 18q, and losses of 6q, 8p, 14q, and 17p occur less frequently (Figure 1) (14,15). Conversely, in NMZL deletion of 7q is absent, while trisomies of chromosomes 3, 18, 7, 12 and del6q are frequent clonal aberrations (Figure 1) (5,6,16).
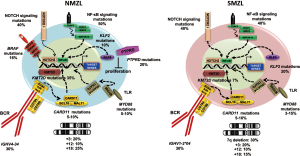
Notably, unlike MALT lymphoma, neither SMZL or NMZL have specific chromosomal translocations involving the IGH locus and pro-survival genes. However, a higher prevalence of KMT2D mutations and loss of function mutations of PTPRD are reported for NMZL (5), and deletions affecting the long arm of chromosome 7 could be of diagnostic use in SMZL (4,17,18). Moreover, increased expression of MNDA has been described as aiding the differential diagnosis between NMZL and follicular lymphoma (19), although the potential of MNDA as a biomarker in NMZL is yet to be determined in independent studies.
SMZL and NMZL
The lack of specific molecular biomarkers hinders precise diagnosis of SMZL or NMZL, particularly in the absence of spleen histology or in case of disseminate disease to the lymph nodes (20). Nevertheless, SMZL is characterized by a sinusoidal dissemination with a frequent nodular pattern of neoplastic cells in the bone marrow that is regarded as a possible hallmark of the disease (21). The course of MZL is usually indolent, although a fraction of patients (15–30%) progresses to a more aggressive disease (22-24). Hence, there is a clinical need for diagnostic and prognostic biomarkers that facilitate diagnosis of the disease and improve the risk stratification of patients.
The assumption that antigen stimulation could play a role in the pathogenesis and progression of MZL is supported by many studies (14,25-27). Evidence of somatic hypermutation (SHM) of the immunoglobulin heavy variable (IGHV) gene and the biased usage of IGHV gene rearrangements, point to antigen selection and expansion (14,25-27). The selective usage of immunoglobulin gene rearrangement IGHV1-2*04 in about 30% of SMZL cases (27,28), and the highly restricted stereotyped configuration of the BCR in about 10% of cases, suggest an antigen stimulation contribution (Figure 1). In NMZL around 30% to 40% of cases present IGHV4-34 usage (28,29), and the mutational status of immunoglobulin gene rearrangements is significantly higher in most cases (Figure 1) (29). However, the specific antigens encountered by B cells expressing IGHV1-2*04 or IGHV4-34 are yet to be determined (30). Infection with the hepatitis C virus (HCV) seems to be related to the use of the IGHV1-69 family in MZL (31-33).
Genomic landscape of SMZL and NMZL
Recent technological advances in genomics and functional genomics have significantly impacted our understanding of the molecular and biological bases of SMZL and NMZL (3-5,7-13,34). An increasingly emerging aspect from genomic studies is that malignant lymphoid cells manipulate signaling pathways that are central to the homeostasis of their normal counterparts. The most frequently mutated genes in SMZL and NMZL are physiologically involved in the normal process of differentiation of mature B cells into MZ cells, and in their trafficking between the functional compartments of lymphoid tissues. MZ development is therefore the major deregulated program in SMZL and NMZL and comprises BCR signaling, Toll-like receptor (TLR) signaling, NF-κB, and NOTCH signaling (3-5,7-13,34).
NF-κB pathway
Both canonical and non-canonical NF-κB pathways play a critical role in the development of MZ B cells. The canonical NF-κB pathway is activated by signaling from several surface receptors including BCR and TLR, and is mainly regulated by its activator IKBKB and inhibitor TNFAIP3. IKBKB encodes a serine kinase that plays an essential role in activation of the canonical NF-κB pathway. The IKBKB protein phosphorylates inhibitors of the NF-κB complex, causing their dissociation and subsequent activation of NF-κB. In SMZL, up to 10% of cases harbor IKBKB hotspot mutations that constitutively activate the protein by changing a highly conserved amino acid of the kinase domain (2). The constitutive activation of NF-κB pathway promotes the growth and survival of tumor cells (2,35). TNFAIP3 encodes a zinc finger protein and ubiquitin-editing enzyme, which has both ubiquitin ligase and deubiquitinase activities and is a principal negative regulator of NF-κB. Frameshift mutations, nonsense mutations or deletions of the entire TNFAIP3 gene locus are observed in about 10% to 15% of SMZL and NMZL (2,3,10). Inactivation of TNFAIP3 causes excessive activation of NF-κB signaling in lymphoma cells, resulting in apoptosis inhibition and oncogenic proliferation (2,36).
Activation of the non-canonical NF-κB pathway by CD40 receptor binding disrupts the TRAF3/MAP3K14-TRAF2/BIRC3 negative regulatory complex. Upon disruption of this complex, the central activating kinase of the pathway, MAP3K14 is released and activated to induce the phosphorylation and proteasomal processing of p100, leading to the formation of p52-RelB dimers. Translocation of p52-RelB to the nucleus enables it to positively regulate the transcription of target genes. Mutations of the TRAF3/MAP3K14-TRAF2/BIRC3 complex are observed in approximately 15% of SMZL, and in 10% of NMZL cases.
The BIRC3 gene encodes a member of the IAP family of proteins. It consists of 3 baculovirus IAP repeats and a C-terminal RING finger domain that confers ubiquitin ligase (E3) activity. Physiologically, BIRC3 catalyzes MAP3K14 protein ubiquitination, resulting in its proteasomal degradation. The RING domain of BIRC3 is inactivated by frameshift or nonsense mutations in 10% of SMZL and 5% of NMZL. Disruption of the RING domain produces a truncated BIRC3 protein that lacks ubiquitin ligase activity, leading to functional stabilization of MAP3K14 and upregulation of NF-κB target genes (2,37).
The TRAF3 gene encodes a member of the TNF receptor associated factor (TRAF) protein family. Members of this family are composed of 3 structural domains: a RING finger in the N-terminal part of the protein, 1-7 TRAF zinc fingers in the central portion and a MATH domain in the C-terminal region; the latter domain is necessary for self-association and receptor interaction. TRAF3 induces MAP3K14 degradation by recruiting it to the BIRC3 ubiquitin ligase complex. In SMZL and NMZL, 5% of cases harbor TRAF3 disrupting mutations in the C-terminal MATH domain that is required for recruiting MAP3K14 to BIRC3, resulting in MAP3K14 stabilization (2,3).
Overall, mutations of positive or negative NF-κB regulators occur in ~40% of SMZL and ~50% of NMZL, indicating the major role of NF-κB genetic lesions in the pathogenesis of SMZL and NMZL (Figure 1) (3-5,9,12,13). NF-κB activation in SMZL and NMZL is also sustained by molecular alterations affecting upstream pathways that are tightly connected to NF-κB activation in B cells, including TLR and BCR signaling.
TLR pathway
TLRs play a critical role in NF-κB pathway activation. Ligation to TLRs induces receptor dimerization and a conformational change that triggers downstream signaling events. Activated TLRs recruit the adaptor protein MYD88 required for propagating the signals from the TLR. MYD88 consists of an N-terminal death domain, an intermediate linker domain, and a C-terminal TIR domain. The death domain allows MYD88 oligomerization and interaction with the respective death domains of IRAKs 1-4 serine-threonine kinases, creating a protein complex called the myddosome. The myddosome complex propagates the signal and activates the NF-κB pathway.
The TIR domain of MYD88 is crucial for signal transduction since it mediates contact with the intracellular TIR domains of TLRs upon signaling activation (38). MYD88-defective subjects are deficient in MZ B cells, suggesting that MYD88 is involved in physiological MZ B cell differentiation (39). MYD88 is affected by somatic mutations mapping within the evolutionarily conserved ββ-loop of the TIR domain in 3% to 15% of SMZL and in up to 10% of NMZL cases (Figure 1) (40-47). These mutations change MYD88 structure, leading to spontaneous homodimerization and recruitment of IRAKs 1-4. The final result is enhanced NF-κB activity (48). The most recurrent mutation in MYD88 is the gain-of-function L265P variant in the toll/interleukin-1 receptor homology (TIR) domain. In SMZL the L265P variant is associated with a superior survival compared with wildtype MYD88 (4).
The functional impact of TLR signaling on the activation and viability of MZ B cells has been demonstrated by their response to inhibitors of IRAK1/4. The TLR-mediated survival effect is blocked by IRAK1/4 inhibitors, suggesting that this pathway may represent a novel therapeutic target and supporting an important role for TLR activation in this disease (48).
BCR pathway
In normal B cells, when the BCR is ligated by an antigen, a signaling cascade is initiated, ultimately resulting in CARD11 phosphorylation and activation. CARD11 protein belongs to the membrane-associated guanylate kinase (MAGUK) family, a class of proteins that functions as molecular scaffolds for the assembly of multiprotein complexes at specialized regions of the plasma membrane. This protein is also a member of the CARD protein family, whose characteristic feature is a caspase-associated recruitment domain (CARD). Upon BCR stimulation, CARD11 acquires an open conformation that allows BCL10 and MALT1 recruitment to activate the CBM (CARD11/BCL10/MALT1) complex, leading to the activation of IKKβ kinase and initiation of NF-κB signaling.
In SMZL and NMZL, CARD11 is mutated in 5% to 10% of cases (Figure 1) (2,5,13,40). CARD11 mutations affect the coiled-coil domain of the protein and disrupt interaction of the coiled-coil domain with the inhibitory domain by promoting spontaneous CARD11 multimerization and association with other CBM complex proteins, resulting in constitutive NF-κB activation (49).
NOTCH pathway
Similar to NF-κB, activation of the NOTCH signaling pathway is a crucial event, in both normal and tumoral MZ B cells. The biochemistry of canonical NOTCH signaling is well established. NOTCH is a family of heterodimeric transmembrane proteins that functions as ligand-activated transcriptional co-factors. Physiologically, differentiation and homing of B cells into the MZ is dependent on NOTCH2 signaling (50-53). NOTCH2 encodes a co-transcription factor that physiologically promotes B cell differentiation into MZ B cells. In mice, Notch2 is critical for promoting MZ B cell differentiation and retention in the splenic MZ (54). The NOTCH2 ligand, DL1, is functionally available to B cells in the spleen (50,51). Upon activation, the cleaved intracellular portion of NOTCH (ICN) translocates into the nucleus, recruits transcriptional co-factors, and modifies expression of target genes. The PEST domain located at the C-terminus of ICN terminates signaling by directing ICN towards proteasomal degradation (52).
NOTCH2 is one of the most frequently mutated genes in SMZL and NMZL, with an occurrence of 10% to 25% and 25%, respectively (3-5,9,11-13). In SMZL, NOTCH2 mutations are mostly represented by monoallelic stop codons or frameshift indels that truncate the PEST domain of ICN2. The loss of the PEST domain prevents proteasomal degradation of ICN2 and stabilizes its expression indicating that NOTCH2 mutations in SMZL and NMZL are gain-of-function events (3). Indeed, genomic studies have identified other regulators of NOTCH signaling that are affected by mutations, including NOTCH1, a paralogue of NOTCH2, which is targeted by similar mutations that truncate the PEST domain and stabilize the NOTCH1 protein (55). NOTCH1 mutations occur in approximately 5% of SMZL cases (3). Negative regulators of NOTCH signaling, including SPEN, DTX1 and MAML2, harbor mutually exclusive mutations in around 40% of SMZL and NMZL cases (Figure 1) (3,13).
KLF2
KLF2 encodes a transcription factor that represses B cell activation and differentiation into MZ B cells. KLF2 is a member of the Krüppel-like factor zinc finger transcription factor family. From a biochemical standpoint, the domains required for its transcriptional activities include three highly conserved zinc fingers at the C-terminus, which allow protein-DNA contact, and two nuclear localization sequences that mediate the localization of KLF2 protein in the nucleus (56-58). In mice, loss of Klf2 expression forces follicular cells of the spleen to gain a MZ-like phenotype and to migrate to the splenic MZ. KLF2 mutations and deletions occur in 30% of SMZL cases and in 20% of NMZL (Figure 1) (5,7,8,34).
KLF2 frameshift and nonsense variants remove the N-terminal zinc finger domain of the protein, including the nuclear localization sequences (4,5,7,8). KLF2 missense substitutions alter amino acids of the nuclear localization sequences, or change highly conserved codons of the first zinc finger domain involved in DNA recognition by KLF2. Consistently, KLF2 mutants translocate from the nucleus into the cytoplasm. Mutated KLF2 inhibits the ability of KLF2 protein to suppress NF-κB induction by upstream signaling pathways, including the BCR and the TLR pathways (7).
The development of splenic MZ B cells requires their differentiation and migration to the marginal sinus in the spleen and their retention there until they differentiate into antibody secreting plasmablasts. Together with other factors, activation of NOTCH2 signaling and depletion of KLF2 enables committed B cells to migrate to the MZ, where they differentiate into MZ B cells and acquire self-renewal and homing capabilities (59-63).
NOTCH2 and KLF2 mutations co-occur in a large fraction of SMZL, suggesting that transcriptional and epigenetic programs required for physiological MZ B cell commitment are constitutively deregulated in SMZL. Deregulated transcription and differentiation are indeed recurrent themes in lymphomagenesis. Although NOTCH2 and KLF2 fulfill the genetic definition of candidate cancer-driver genes, they are not biologically validated as oncogenes or tumor-suppressors in SMZL. Transgenic mice engineered to harbor either constitutive NOTCH2 activation or lacking KLF2 in B cells display splenomegaly due to the expansion of the MZ B cell compartment, but as yet, none have been reported to have a lymphoma phenotype (59-62).
The contribution of NOTCH2 and KLF2 aberrations to SMZL pathogenesis is still unclear. Some crucial questions need to be addressed by further functional studies in SMZL; firstly, there is no evidence of NOTCH2 and KLF2 being either independent or concomitant drivers in SMZL. Secondly, comprehensive knowledge about the genes targeted by NOTCH2 and KLF2 in MZ B cells is lacking, and thirdly, the consequences of NOTCH2 and KLF2 mutations on the signaling/epigenetic programs of SMZL are not well understood.
PTPRD
PTPRD encodes the receptor-type-protein-tyrosine-phosphatase δ, a tumor suppressor gene involved in cell growth regulation and expressed in naïve, germinal center, and MZ B cells (5). PTPRD is physiologically involved in several cellular programs, including proliferation and downregulation of cytokine signaling via dephosphorylation of STAT3 at tyrosine 705 (Y705). In NMZL PTPRD mutations occur in ~20% of cases; in contrast PTPRD mutations are absent in SMZL cases (Figure 1) (5). PTPRD genetic lesions in NMZL include chromosomal deletions or missense mutations, which cause loss of the entire PTPRD gene or damage the protein tyrosine phosphatase function, respectively. In fact, NMZL cases harboring PTPRD lesions show significant enrichment of cell cycle genes compared to PTPRD wild type cases, including higher expression of Ki-67, indicating that PTPRD is involved in cell cycle regulation in normal and malignant B cells (5). Aberrations affecting PTPRD might therefore represent a promising biomarker for NMZL, although their prevalence has to be confirmed by additional independent studies.
Epigenetic regulator genes
Genomic studies have also identified mutations in several epigenetic regulators that occur in about 40% of SMZL and NMZL cases (Figure 1). Although the functional consequences of these mutations have not yet been determined, the genes affected include KMT2D, ARID1A, EP300, CREBBP, SIN3A and TBL1XR1 (3,5).
BRAF
Mutations of BRAF have recently been reported in NMZL (16%, 4 out of 25 cases) and might be exclusive for NMZL across MZL subtypes, suggesting a diagnostic potential of BRAF in NMZL (Figure 1) (34). However, since BRAF mutations in NMZL have only been reported in a single study, their prevalence should be validated in larger, independent series.
Transcriptomic profiles
The gene expression profiles of SMZL and NMZL have been well characterized (64-69). Overexpression of members of BCR-TLR-NF-κB, PI3K-AKT, JAK-STAT and RHO GTPases signaling cascades, together with immune system and proliferation genes, as well as markers of normal MZ B cells, has been consistently and independently observed (64-69). The top commonly up-regulated transcripts encode B cell kinases downstream of the BCR and pro-survival genes such as SYK, BIRC3, BTK, CD40, TLR9, CD79A-B, PLCγ2, CARD11, RHOH, IL6 and TCL1A (Table 1). The expression patterns of SMZL and NMZL thus reveal the activation of pro-survival signatures that might contribute to the development or progression of MZL. Further, the kinases and survival genes identified in these studies provide a rationale for the use of small molecules to target therapeutic vulnerabilities.
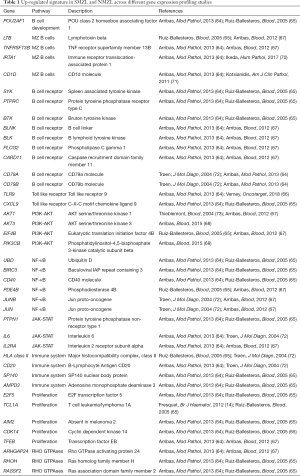
Full table
Methylation patterns
Genome-wide epigenetic profiling of SMZL has identified aberrant promoter DNA methylation as a mechanism of deregulation that affects crucial pathways in the disease. A high degree of promoter DNA methylation is observed in 25% of SMZL cases and these patients show poorer overall survival and enriched IGHV1-02*04 usage, NOTCH2 mutations, 7q31-32 loss and histological transformation to aggressive lymphoma. This high methylation SMZL subset might be useful in risk stratification of patients (68).
Aberrant methylation seems to contribute to the high expression of pro-survival genes (TCL1B, BCL2A1, FGF1), BCR and NF-κB signaling members (CD79B, CARD11, PIK3CB), PRC2-complex genes (EZH2, EED, SUZ12), and genes involved in JAK-STAT and PI3K/AKT signaling pathways. Several tumor suppressor genes appear highly methylated, such as KLF4, CDKN1C, CDKN2A/B/D, CDH1-2, WT1, RARB and GATA4. Thus, aberrant methylation could also play a role in the pathogenesis and/or progression of SMZL (68,69). In accordance with this, genomic variants affecting epigenetic regulators have been identified in SMZL and these might be partially responsible for the aberrant methylation and subsequent development of the disease.
At the moment of writing this review there were no studies characterizing the genome-wide methylation profiles of NMZL, although promoter methylation might collaborate with loss-of-function mutations in repressing the tumor suppressor gene PTPRD (5).
miRNA profiles
In the last few years, a number of studies have investigated the role of miRNAs in MZL; only a few of these determined the miRNA profiles of NMZL (64,67,74,75). On the contrary, the miRNome of SMZL has been more extensively investigated in the context of the 7q LOH (15,76,77) and also in comparisons of SMZL to non-tumoral tissues (64,76,78), or to other B cell lymphomas (64,74).
The frequency of 7q loss in SMZL (~30%) is one of the highest for chromosomal aberrations in lymphomas. In the aforementioned studies focused on miRNAs located on 7q, the miR-29a/miR-29b-1 cluster was proposed as a target of the 7q LOH, involved in up-regulating the TCL1A oncogene (77), and miRNAs located within the minimal deleted region exhibited decreased expression in SMZL patients harboring deletion of 7q (15). Despite the prevalence of HCV infection reported in SMZL (31-33), no specific miRNA profile was associated with HCV in SMZL, although repression of miR-26b was observed in the HCV+ cases (78). In 2015, a comprehensive study compared miRNA expression patterns between indolent NMZL, transformed NMZL and diffuse large B cell lymphoma (DLBCL). Interestingly there were no significant differences between the miRNA profiles of indolent and transformed NMZL, although the authors identified decreased expression of miRNAs in transformed NMZL compared to DLBCL, including members of the miR-183/96/182 polycistron (75). Most recently reviewed was the involvement of miRNAs in the pathogenesis of SMZL, where miRNAs located at 7q or those targeting the 7q31 gene CAV1, were proposed as promising candidates for further investigation (79).
Only a few miRNAs have been found consistently de-regulated across these studies and different factors likely contribute to this variability, including the high degree of heterogeneity in SMZL and differences in experimental designs. Additionally, we are still far from fully understanding the biology of non-coding RNAs. In the following paragraphs we will discuss in more depth the miRNAs that have been identified in various independent studies.
High expression levels of miR-155 have been reported in both SMZL and NMZL (64,74,76,78), while overexpression of miR-21 is found in SMZL compared to reactive spleen and is also associated with aggressive SMZL (64,76,78). The overexpression of miR-21 and miR-155 seems to contribute to the activation of survival signaling pathways in B cell lymphoma. An elevated expression of miR-21 has been extensively reported in hematopoietic disorders (80-84). Mechanistically, miR-21 downmodulates tumor suppressor genes, including PDCD4 and PTEN, which negatively regulate MAPK and PI3K-AKT pathways respectively (80,84). In a similar manner to miR-21, miR-155 is strongly up-regulated across B cell lymphoma subtypes (84-89). miR-155 is one of the most well studied miRNAs in B cells and plays a role in the regulation of many crucial processes. For example, miR-155 is involved in B cell differentiation (85) and plays a fundamental role in the regulation of BCR signaling. Ligand engagement by BCR induces miR-155 expression (90), and directly targets negative regulators of downstream BCR signaling, such as SHIP1 and FOXO3 (88,91,92). BCR signaling leads to activation of NF-κB, which then induces expression of both miR-155 and miR-21. This positive feedback loop likely sustains high levels of these miRNAs (93,94).
Conversely, miRNAs with potential tumor suppressor capabilities have been reported in SMZL. Mir-136 is repressed in SMZL compared to non-tumoral spleen (64) but exhibits higher expression when compared to NMZL and MALT lymphoma (74), suggesting a specific expression pattern of miR-136 in MZL subtypes. Although the mechanistic role of miR-136 has not yet been characterized in lymphoma, in solid tumors miR-136 acts as a tumor suppressor that regulates proliferation, migration and invasion (95), and targets BCL2 to promote apoptosis (96). Experiments in rats have shown that miR-136 regulates the activation of NF-κB, since its forced expression induces secretion of pro-inflammatory cytokines and inhibits the expression of the NF-κB negative regulator TNFAIP3 (97).
The expression levels of miR-127 are lower in SMZL compared to non-tumoral spleen (64), and in accordance with the expansion of the MZ in SMZL, miR-127 is not expressed in normal memory B cells (98). Indeed, miR-127 targets crucial regulators of B cell development, including BCL6 and CD10 (99), whose expression is absent in SMZL tumors.
Very few miRNAs are differentially expressed in NMZL. Compared to non-tumoral tissue, mir-370 and miR-513 are downregulated in NMZL and conversely, miR-221 and miR-223 appear up-regulated in NMZL when compared to other B cell lymphomas (67,74). Lack of epigenetic regulation resulting from the loss of miRNAs may play a role in immune escape of tumor cells. Disruption of miR-513 induces PD-L1 protein expression, and transfection of miR-513 into Jurkat cells eradicates PD-L1 expression and inhibits apoptosis (100). Secretion of pro-inflammatory cytokines, including IL6 and IL1β, is abolished by miR-370 (101). Downmodulation of miR-370 in DLBCL increases the expression of members of BCR and PI3K signaling cascades and its expression is associated with sensitivity to rituximab and doxorubicin (102). Furthermore, aberrant homing and differentiation of tumor cells may also be partly due to epigenetic de-regulation. In this context, miR-221 and miR-223 are thought to have roles in B cell differentiation and homing (103,104); NMZL clinical specimens express genes related to the normal MZ B cell signature and thus lack expression of germinal center markers such as LMO2 and CD10, which are targeted by miR-223 and miR-221 (67). Additionally, studies in DLBCL have reported high expression of miR-223 compared to non-tumoral lymph nodes (105), and miR-221 is overexpressed in activated B cell-like compared to the germinal center B cell-like DLBCL subtypes (85). Therefore, profiles of miR-221 and miR-223 in B cell lymphoma are consistent with the notion that both miRNAs might contribute to the specific signature of post germinal center B cells. Lastly but of clinical interest, miR-223 may represent a novel prognostic biomarker since longer survival of DLBCL (105) or CLL (106) patients correlates with high expression of miR-223.
Molecular pathogenesis of SMZL and NMZL
The genomic and transcriptomic landscapes of SMZL and NMZL indicate a role for NF-κB activation in the pathogenesis of MZL (20,64,67,107). Not surprisingly, signals from NF-κB are necessary for normal establishment and maintenance of the MZ (108). Together with the high prevalence of molecular lesions targeting NF-κB pathway components in ~40-50% of SMZL and NMZL patients, these findings underscore NF-κB pathway activation as a hallmark of these MZL subtypes (2-6,10,12,13). Overexpression of genes involved in NF-κB signaling has been reported in a number of genomic studies (64,65), including upstream signals from BCR (SYK, BTK, CD79A-B), TLR (TLR9, CXCL9) and CD40, as well as a number of NF-κB related transcripts (UBD, BIRC3, TRAF3, IL6, JUN, FOS). Supporting the transcriptional activation of the NF-κB signaling cascade, approximately half of SMZL patients show nuclear expression of p50 and/or p52 (2).
The up-regulation of BCR/TLR/NF-κB in MZL might be the consequence of concomitant genomic and epigenetic events. Somatic mutations in key regulators of the NF-κB pathway, aberrant DNA methylation and de-regulated miRNAs likely function in concert to activate the NF-κB cascade. In such a scenario, mutations of NF-κB pathway members and upstream signaling components (TNFAIP3, BIRC3, CARD11, MAP3K14, TRAF3, MYD88), KLF2 genomic lesions, low methylation of promoters regulating BCR/TLR/NF-κB members and aberrant miRNA expression (overexpression of miR-155, repression of miR-27b, miR-370 and miR-377), lead to constitutively active signaling downstream of the BCR.
Although NF-κB is probably the most relevant survival pathway activated in SMZL and NMZL, other signaling cascades have been reported with enhanced activity, which might have therapeutic implications. Members of the PI3K/AKT and JAK/STAT signaling pathways are unmethylated and highly expressed in SMZL (68) and in NMZL (67). In SMZL up-regulation of miR-21, which targets PTEN, might work together with the low methylation levels of PI3K signaling genes (AKT1, AKT3, PIK3CB and CXCR4) to determine the overexpression and activation of the pathway.
A consequence of continuous NF-κB signals is the secretion of pro-inflammatory cytokines, such as IL6 and IL2 (109,110), which induces the NF-κB transcription machinery resulting in a positive feedback loop that can over activate NF-κB signaling (111). Release of IL6 and/or IL2 also initiates a cascade of kinases whose phosphorylating action triggers the JAK/STAT pathway. Since members of the JAK/STAT pathway are reported to be unmethylated in SMZL (68), and upregulated in both SMZL and NMZL (67), it is possible that these perturbations establish a cancer-related pro-inflammatory circuit (111).
Biomarkers for diagnosis/prognosis
Diagnosis, prognosis and development of new therapeutic approaches might take advantage of molecular lesions and potential biomarkers so far identified in MZL. Loss of 7q or TNFAIP3, together with the presence of trisomies 3 and 18 indicate a diagnosis of MZL. Mutations of NF-κB members might not have a diagnostic value in SMZL or NMZL since they are not specific for these entities within the spectrum of B cell lymphomas (107). Mutations affecting NOTCH2 suggest a NMZL or SMZL diagnosis with the possible exclusion of MALT lymphoma (19,20). Nevertheless, these molecular biomarkers might not be adequately sensitive and are not specific for SMZL or NMZL.
A number of retrospective studies have described biomarkers of prognosis in NMZL, although they should be considered with precaution due to the limited number of patients and different therapeutic approaches (112). Despite these caveats, molecular lesions with a high prevalence in SMZL show some promise as prognostic biomarkers, including KLF2 and NOTCH2 mutations that are linked to poorer outcome in SMZL (4,8,13).
Future perspectives
The classification of lymphoma by the WHO recognized SMZL and NMZL as distinct entities with specific phenotypical, morphological and clinical features. Nonetheless, similar incidences and common aspects in the biology of SMZL and NMZL are consistent with a common origin in MZ B cells. Many studies have improved diagnosis and shed light on the biology of these tumors however, the causes and pathogenic mechanisms of SMZL and NMZL are yet to be clarified. The genetic landscapes of SMZL and NMZL exhibit common genomic aberrations of crucial players in MZL that are mostly regulated upon NF-κB activation, such as NOTCH2 and KLF2. These common lesions present opportunities for novel therapeutic approaches that might benefit both SMZL and NMZL patients. Additionally, these mutated genes might be promising biomarkers for SMZL and/or NMZL, including NOTCH2, KLF2, PTPRD and BRAF, although their diagnostic and prognostic relevance requires further investigations.
At present, current therapies for NMZL patients are still based on immunochemotherapy, while many SMZL cases still undergo splenectomy or rituximab monotherapy as a valid treatment. The expectation is that novel personalized medicine approaches will provide molecularly targeted therapies that match the unique genetic, epigenetic and clonal composition of each individual tumor.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Francesco Bertoni, Thomas Habermann, Davide Rossi, Emanuele Zucca) for the series “Marginal Zone Lymphomas” published in Annals of Lymphoma. The article has undergone external peer review.
Conflicts of Interest: All three authors have completed the ICMJE uniform disclosure form (http://dx.doi.org/10.21037/aol-20-38). The series “Marginal Zone Lymphomas” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Swerdlow S, Campo E, Harris N, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Geneva: IARC Press, 2017.
- Rossi D, Deaglio S, Dominguez-Sola D, et al. Alteration of BIRC3 and multiple other NF-κB pathway genes in splenic marginal zone lymphoma. Blood 2011;118:4930-4. [Crossref] [PubMed]
- Rossi D, Trifonov V, Fangazio M, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med 2012;209:1537-51. [Crossref] [PubMed]
- Parry M, Rose-Zerilli MJ, Ljungström V, et al. Genetics and prognostication in splenic marginal zone lymphoma: revelations from deep sequencing. Clin Cancer Res 2015;21:4174-83. [Crossref] [PubMed]
- Spina V, Khiabanian H, Messina M, et al. The genetics of nodal marginal zone lymphoma. Blood 2016;128:1362-73. [Crossref] [PubMed]
- Rinaldi A, Mian M, Chigrinova E, et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood 2011;117:1595-604. [Crossref] [PubMed]
- Clipson A, Wang M, de Leval L, et al. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia 2015;29:1177-85. [Crossref] [PubMed]
- Piva R, Deaglio S, Famà R, et al. The Krüppel-like factor 2 transcription factor gene is recurrently mutated in splenic marginal zone lymphoma. Leukemia 2015;29:503-7. [Crossref] [PubMed]
- Bruscaggin A, Monti S, Arcaini L, et al. Molecular lesions of signalling pathway genes in clonal B-cell lymphocytosis with marginal zone features. Br J Haematol 2014;167:718-20. [Crossref] [PubMed]
- Novak U, Rinaldi A, Kwee I, et al. The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood 2009;113:4918-21. [Crossref] [PubMed]
- Parry M, Rose-Zerilli MJ, et al. Whole exome sequencing identifies novel recurrently mutated genes in patients with splenic marginal zone lymphoma. PLoS One 2013;8:e83244 [Crossref] [PubMed]
- Martínez N, Almaraz C, Vaqué JP, et al. Whole-exome sequencing in splenic marginal zone lymphoma reveals mutations in genes involved in marginal zone differentiation. Leukemia 2014;28:1334-40. [Crossref] [PubMed]
- Kiel MJ, Velusamy T, Betz BL, et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med 2012;209:1553-65. [Crossref] [PubMed]
- Fresquet V, Robles EF, Parker A, et al. High-throughput sequencing analysis of the chromosome 7q32 deletion reveals IRF5 as a potential tumour suppressor in splenic marginal-zone lymphoma. Br J Haematol 2012;158:712-26. [Crossref] [PubMed]
- Watkins AJ, Hamoudi RA, Zeng N, et al. An integrated genomic and expression analysis of 7q deletion in splenic marginal zone lymphoma. PLoS One 2012;7:e44997 [Crossref] [PubMed]
- van den Brand M, van Krieken JH. Recognizing nodal marginal zone lymphoma: recent advances and pitfalls. A systematic review. Haematologica 2013;98:1003-13. [Crossref] [PubMed]
- Algara P, Mateo MS, Sanchez-Beato M, et al. Analysis of the IgV(H) somatic mutations in splenic marginal zone lymphoma defines a group of unmutated cases with frequent 7q deletion and adverse clinical course. Blood 2002;99:1299-304. [Crossref] [PubMed]
- Watkins AJ, Huang Y, Ye H, et al. Splenic marginal zone lymphoma: characterization of 7q deletion and its value in diagnosis. J Pathol 2010;220:461-74. [Crossref] [PubMed]
- Kanellis G, Roncador G, Arribas A, et al. Identification of MNDA as a new marker for nodal marginal zone lymphoma. Leukemia 2009;23:1847-57. [Crossref] [PubMed]
- Bertoni F, Rossi D, Zucca E. Recent advances in understanding the biology of marginal zone lymphoma. F1000Res 2018;7:406. [Crossref] [PubMed]
- Arcaini L, Varettoni M, Boveri E, et al. Distinctive clinical and histological features of Waldenström's macroglobulinemia and splenic marginal zone lymphoma. Clin Lymphoma Myeloma Leuk 2011;11:103-5. [Crossref] [PubMed]
- Montalbán C, Abraira V, Arcaini L, et al. Risk stratification for splenic marginal zone lymphoma based on haemoglobin concentration, platelet count, high lactate dehydrogenase level and extrahilar lymphadenopathy: development and validation on 593 cases. Br J Haematol 2012;159:164-71. [Crossref] [PubMed]
- Oh SY, Ryoo BY, Kim WS, et al. Nodal marginal zone B-cell lymphoma: Analysis of 36 cases. Clinical presentation and treatment outcomes of nodal marginal zone B-cell lymphoma. Ann Hematol 2006;85:781-6. [Crossref] [PubMed]
- Arcaini L, Lazzarino M, Colombo N, et al. Splenic marginal zone lymphoma: a prognostic model for clinical use. Blood 2006;107:4643-9. [Crossref] [PubMed]
- Conconi A, Bertoni F, Pedrinis E, et al. Nodal marginal zone B-cell lymphomas may arise from different subsets of marginal zone B lymphocytes. Blood 2001;98:781-6. [Crossref] [PubMed]
- Tierens A, Delabie J, Pittaluga S, et al. Mutation analysis of the rearranged immunoglobulin heavy chain genes of marginal zone cell lymphomas indicates an origin from different marginal zone B lymphocyte subsets. Blood 1998;91:2381-6. [Crossref] [PubMed]
- Rinaldi A, Forconi F, Arcaini L, et al. Immunogenetics features and genomic lesions in splenic marginal zone lymphoma. Br J Haematol 2010;151:435-9. [Crossref] [PubMed]
- Zibellini S, Capello D, Forconi F, et al. Stereotyped patterns of B-cell receptor in splenic marginal zone lymphoma. Haematologica 2010;95:1792-6. [Crossref] [PubMed]
- Traverse-Glehen A, Davi F, Ben Simon E, et al. Analysis of VH genes in marginal zone lymphoma reveals marked heterogeneity between splenic and nodal tumors and suggests the existence of clonal selection. Haematologica 2005;90:470-8. [PubMed]
- Bikos V, Karypidou M, Stalika E, et al. An Immunogenetic Signature of Ongoing Antigen Interactions in Splenic Marginal Zone Lymphoma Expressing IGHV1-2*04 Receptors. Clin Cancer Res 2016;22:2032-40. [Crossref] [PubMed]
- Couronné L, Bachy E, Roulland S, et al. From hepatitis C virus infection to B-cell lymphoma. Ann Oncol 2018;29:92-100. [Crossref] [PubMed]
- Vannata B, Zucca E. Hepatitis C virus-associated B-cell non-Hodgkin lymphomas. Hematology Am Soc Hematol Educ Program 2014;2014:590-8. [Crossref] [PubMed]
- Marasca R, Vaccari P, Luppi M, et al. Immunoglobulin gene mutations and frequent use of VH1-69 and VH4-34 segments in hepatitis C virus-positive and hepatitis C virus-negative nodal marginal zone B-cell lymphoma. Am J Pathol 2001;159:253-61. [Crossref] [PubMed]
- Pillonel V, Juskevicius D, Ng CKY, et al. High-throughput sequencing of nodal marginal zone lymphomas identifies recurrent BRAF mutations. Leukemia 2018;32:2412-26. [Crossref] [PubMed]
- Kai X, Chellappa V, Donado C, et al. IκB kinase β (IKBKB) mutations in lymphomas that constitutively activate canonical nuclear factor κB (NFκB) signaling. J Biol Chem 2014;289:26960-72. [Crossref] [PubMed]
- Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009;459:717-21. [Crossref] [PubMed]
- Rahal R, Frick M, Romero R, et al. Pharmacological and genomic profiling identifies NF-κB-targeted treatment strategies for mantle cell lymphoma. Nat Med 2014;20:87-92. [Crossref] [PubMed]
- Rossi D. Role of MYD88 in lymphoplasmacytic lymphoma diagnosis and pathogenesis. Hematology Am Soc Hematol Educ Program 2014;2014:113-8. [Crossref] [PubMed]
- Wang JQ, Jeelall YS, Beutler B, et al. Consequences of the recurrent MYD88 (L265P) somatic mutation for B cell tolerance. J Exp Med 2014;211:413-26. [Crossref] [PubMed]
- Yan Q, Huang Y, Watkins AJ, et al. BCR and TLR signaling pathways are recurrently targeted by genetic changes in splenic marginal zone lymphomas. Haematologica 2012;97:595-8. [Crossref] [PubMed]
- Gachard N, Parrens M, Soubeyran I, et al. IGHV gene features and MYD88 L265P mutation separate the three marginal zone lymphoma entities and Waldenström macroglobulinemia/lymphoplasmacytic lymphomas. Leukemia 2013;27:183-9. [Crossref] [PubMed]
- Xu L, Hunter ZR, Yang G, et al. MYD88 L265P in Waldenström macroglobulinemia, immunoglobulin M monoclonal gammopathy, and other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. Blood 2013;121:2051-8. [Crossref] [PubMed]
- Trøen G, Warsame A, Delabie J. CD79B and MYD88 mutations in splenic marginal zone lymphoma. ISRN Oncol 2013;2013:252318 [Crossref] [PubMed]
- Varettoni M, Arcaini L, Zibellini S, et al. Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenstrom's macroglobulinemia and related lymphoid neoplasms. Blood 2013;121:2522-8. [Crossref] [PubMed]
- Hamadeh F, MacNamara SP, Aguilera NS, et al. MYD88 L265P mutation analysis helps define nodal lymphoplasmacytic lymphoma. Mod Pathol 2015;28:564-74. [Crossref] [PubMed]
- Martinez-Lopez A, Curiel-Olmo S, Mollejo M, et al. MYD88 (L265P) somatic mutation in marginal zone B-cell lymphoma. Am J Surg Pathol 2015;39:644-51. [Crossref] [PubMed]
- Peveling-Oberhag J, Wolters F, Döring C, et al. Whole exome sequencing of microdissected splenic marginal zone lymphoma: a study to discover novel tumor-specific mutations. BMC Cancer 2015;15:773. [Crossref] [PubMed]
- Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011;470:115-9. [Crossref] [PubMed]
- Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 2008;319:1676-9. [Crossref] [PubMed]
- Descatoire M, Weller S, Irtan S, et al. Identification of a human splenic marginal zone B cell precursor with NOTCH2-dependent differentiation properties. J Exp Med 2014;211:987-1000. [Crossref] [PubMed]
- Hozumi K, Negishi N, Suzuki D, et al. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol 2004;5:638-44. [Crossref] [PubMed]
- Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity 2010;32:14-27. [Crossref] [PubMed]
- Witt CM, Won WJ, Hurez V, et al. Notch2 haploinsufficiency results in diminished B1 B cells and a severe reduction in marginal zone B cells. J Immunol 2003;171:2783-8. [Crossref] [PubMed]
- Moran ST, Cariappa A, Liu H, et al. Synergism between NF-kappa B1/p50 and Notch2 during the development of marginal zone B lymphocytes. J Immunol 2007;179:195-200. [Crossref] [PubMed]
- Arruga F, Gizdic B, Serra S, et al. Functional impact of NOTCH1 mutations in chronic lymphocytic leukemia. Leukemia 2014;28:1060-70. [Crossref] [PubMed]
- Shields JM, Yang VW. Two potent nuclear localization signals in the gut-enriched Krüppel-like factor define a subfamily of closely related Krüppel proteins. J Biol Chem 1997;272:18504-7. [Crossref] [PubMed]
- Kozyrev SV, Hansen LL, Poltaraus AB, et al. Structure of the human CpG-island-containing lung Kruppel-like factor (LKLF) gene and its location in chromosome 19p13.11-13 locus. FEBS Lett 1999;448:149-52. [Crossref] [PubMed]
- Wani MA, Conkright MD, Jeffries S, et al. cDNA isolation, genomic structure, regulation, and chromosomal localization of human lung Kruppel-like factor. Genomics 1999;60:78-86. [Crossref] [PubMed]
- Hoek KL, Gordy LE, Collinset PL. al. Follicular B cell trafficking within the spleen actively restricts humoral immune responses. Immunity 2010;33:254-65. [Crossref] [PubMed]
- Hampel F, Ehrenberg S, Hojer C, et al. CD19-independent instruction of murine marginal zone B-cell development by constitutive Notch2 signaling. Blood 2011;118:6321-31. [Crossref] [PubMed]
- Hart GT, Wang X, Hogquistet KA, et al. Krüppel-like factor 2 (KLF2) regulates B-cell reactivity, subset differentiation, and trafficking molecule expression. Proc Natl Acad Sci U S A 2011;108:716-21. [Crossref] [PubMed]
- Winkelmann R, Sandrock L, Porstneret M, et al. B cell homeostasis and plasma cell homing controlled by Krüppel-like factor 2. Proc Natl Acad Sci U S A 2011;108:710-5. [Crossref] [PubMed]
- Valls E, Lobry C, Geng H, et al. BCL6 antagonizes NOTCH2 to maintain survival of human follicular lymphoma cells. Cancer Discov 2017;7:506-21. [Crossref] [PubMed]
- Arribas AJ, Gómez-Abad C, Sánchez-Beato M, et al. Splenic marginal zone lymphoma: comprehensive analysis of gene expression and miRNA profiling. Mod Pathol 2013;26:889-901. [Crossref] [PubMed]
- Ruiz-Ballesteros E, Mollejo M, Rodriguez A, et al. Splenic marginal zone lymphoma: proposal of new diagnostic and prognostic markers identified after tissue and cDNA microarray analysis. Blood 2005;106:1831-8. [Crossref] [PubMed]
- Verney A, Traverse-Glehen A, Callet-Bauchu E, et al. Toll-like receptor expression and function differ between splenic marginal zone B cell lymphoma and splenic diffuse red pulp B cell lymphoma. Oncotarget 2018;9:23589-98. [Crossref] [PubMed]
- Arribas AJ, Campos-Martín Y, Gómez-Abad C, et al. Nodal marginal zone lymphoma: gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood 2012;119:e9-21. [Crossref] [PubMed]
- Arribas AJ, Rinaldi A, Mensah AA, et al. DNA methylation profiling identifies two splenic marginal zone lymphoma subgroups with different clinical and genetic features. Blood 2015;125:1922-31. [Crossref] [PubMed]
- Arribas AJ, Bertoni F. Methylation patterns in marginal zone lymphoma. Best Pract Res Clin Haematol 2017;30:24-31. [Crossref] [PubMed]
- Ikeda JI. Immunohistochemical analysis of the novel marginal zone B-cell marker IRTA1 in malignant lymphoma. Hum Pathol 2017;59:70-9. [Crossref] [PubMed]
- Kotsianidis I, Nakou E, Spanoudakis E, et al. The diagnostic value of CD1d expression in a large cohort of patients with B-cell chronic lymphoproliferative disorders. Am J Clin Pathol 2011;136:400-8. [Crossref] [PubMed]
- Trøen G, Nygaard V, Jenssen TK, et al. Constitutive expression of the AP-1 transcription factors c-jun, junD, junB, and c-fos and the marginal zone B-cell transcription factor Notch2 in splenic marginal zone lymphoma. J Mol Diagn 2004;6:297-307. [Crossref] [PubMed]
- Thieblemont C, Nasser V, Felman P, et al. Small lymphocytic lymphoma, marginal zone B-cell lymphoma, and mantle cell lymphoma exhibit distinct gene-expression profiles allowing molecular diagnosis. Blood 2004;103:2727-37. [Crossref] [PubMed]
- Di Lisio L, Martinez N, Montes-Moreno S, et al. The role of miRNAs in the pathogenesis and diagnosis of B-cell lymphomas. Blood 2012;120:1782-90. [Crossref] [PubMed]
- Gebauer N, Thorns C, Bernard V, et al. MicroRNA profiling of low-grade and transformed nodal marginal zone lymphoma reveals a similar signature pattern distinct from diffuse large B cell lymphoma. Acta Haematol 2015;133:214-20. [Crossref] [PubMed]
- Bouteloup M, Verney A, Rachinel N, et al. MicroRNA expression profile in splenic marginal zone lymphoma. Br J Haematol 2012;156:279-81. [Crossref] [PubMed]
- Ruiz-Ballesteros E, Mollejo M, Mateo M, et al. MicroRNA losses in the frequently deleted region of 7q in SMZL. Leukemia 2007;21:2547-9. [Crossref] [PubMed]
- Peveling-Oberhag J, Crisman G, Schmidt A, et al. Dysregulation of global microRNA expression in splenic marginal zone lymphoma and influence of chronic hepatitis C virus infection. Leukemia 2012;26:1654-62. [Crossref] [PubMed]
- Robinson JE, Cutucache CE. Deciphering splenic marginal zone lymphoma pathogenesis: the proposed role of microRNA. Oncotarget 2018;9:30005-22. [Crossref] [PubMed]
- Yamanaka Y, Tagawa H, Takahashi N, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood 2009;114:3265-75. [Crossref] [PubMed]
- Cui Q, Vari F, Cristino AS, et al. Circulating cell-free miR-494 and miR-21 are disease response biomarkers associated with interim-positron emission tomography response in patients with diffuse large B-cell lymphoma. Oncotarget 2018;9:34644-57. [Crossref] [PubMed]
- Li J, Fu R, Yang L, et al. miR-21 expression predicts prognosis in diffuse large B-cell lymphoma. Int J Clin Exp Pathol 2015;8:15019-24. [PubMed]
- Hershkovitz-Rokah O, Geva P, Salmon-Divon M, et al. Network analysis of microRNAs, genes and their regulation in diffuse and follicular B-cell lymphomas. Oncotarget 2018;9:7928-41. [Crossref] [PubMed]
- Rossi S, Shimizu M, Barbarotto E, et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood 2010;116:945-52. [Crossref] [PubMed]
- Lawrie CH, Soneji S, Marafioti T, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer 2007;121:1156-61. [Crossref] [PubMed]
- Di Lisio L, Gómez-López G, Sánchez-Beato M, et al. Mantle cell lymphoma: transcriptional regulation by microRNAs. Leukemia 2010;24:1335-42. [Crossref] [PubMed]
- Roehle A, Hoefig KP, Repsilber D, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol 2008;142:732-44. [Crossref] [PubMed]
- Cui B, Chen L, Zhang S, et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood 2014;124:546-54. [Crossref] [PubMed]
- Vargova K, Curik N, Burda P, et al. MYB transcriptionally regulates the miR-155 host gene in chronic lymphocytic leukemia. Blood 2011;117:3816-25. [Crossref] [PubMed]
- Vigorito E, Perks KL, Abreu-Goodger C, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007;27:847-59. [Crossref] [PubMed]
- Thompson MG, Larson M, Vidrine A, et al. FOXO3-NF-κB RelA protein complexes reduce proinflammatory cell signaling and function. J Immunol 2015;195:5637-47. [Crossref] [PubMed]
- Pedersen IM, Otero D, Kao E, Miletic AV, et al. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol Med 2009;1:288-95. [Crossref] [PubMed]
- Bai H, Wei J, Deng C, et al. MicroRNA-21 regulates the sensitivity of diffuse large B-cell lymphoma cells to the CHOP chemotherapy regimen. Int J Hematol 2013;97:223-31. [Crossref] [PubMed]
- Thompson RC, Herscovitch M, Zhao I, et al. NF-kappaB down-regulates expression of the B-lymphoma marker CD10 through a miR-155/PU.1 pathway. J Biol Chem 2011;286:1675-82. [Crossref] [PubMed]
- Chu Y, Hu X, Wang G, et al. Downregulation of miR-136 promotes the progression of osteosarcoma and is associated with the prognosis of patients with osteosarcoma. Oncol Lett 2019;17:5210-8. [Crossref] [PubMed]
- Yu L, Zhou GQ, Li DC. MiR-136 triggers apoptosis in human gastric cancer cells by targeting AEG-1 and BCL2. Eur Rev Med Pharmacol Sci 2018;22:7251-6. [PubMed]
- Deng G, Gao Y, Cen Z, et al. miR-136-5p regulates the inflammatory response by targeting the IKKbeta/NF-kappaB/A20 pathway after spinal cord injury. Cell Physiol Biochem 2018;50:512-24. [Crossref] [PubMed]
- Leucci E, Onnis A, Cocco M, et al. B-cell differentiation in EBV-positive Burkitt lymphoma is impaired at posttranscriptional level by miRNA-altered expression. Int J Cancer 2010;126:1316-26. [Crossref] [PubMed]
- Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006;9:435-43. [Crossref] [PubMed]
- Gong AY, Zhou R, Hu G, et al. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol 2009;182:1325-33. [Crossref] [PubMed]
- Tian D, Sha Y, Lu JM, et al. MiR-370 inhibits vascular inflammation and oxidative stress triggered by oxidized low-density lipoprotein through targeting TLR4. J Cell Biochem 2018;119:6231-7. [Crossref] [PubMed]
- Leivonen SK, Icay K, Jäntti K, et al. MicroRNAs regulate key cell survival pathways and mediate chemosensitivity during progression of diffuse large B-cell lymphoma. Blood Cancer J 2017;7:654. [Crossref] [PubMed]
- Zhang J, Jima DD, Jacobs C, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood 2009;113:4586-94. [Crossref] [PubMed]
- Knoll M, Simmons S, Bouquet C. miR-221 redirects precursor B cells to the BM and regulates their residence. Eur J Immunol 2013;43:2497-506. [Crossref] [PubMed]
- Yao XX, Wang JF, Wang YH, et al. Expression of microRNA-223 and its clinicopathologic correlation in diffuse large B-cell lymphoma. Zhonghua Bing Li Xue Za Zhi 2012;41:366-70. [PubMed]
- Zhou K, Yi S, Yu Z, et al. MicroRNA-223 expression is uniformly down-regulated in B cell lymphoproliferative disorders and is associated with poor survival in patients with chronic lymphocytic leukemia. Leuk Lymphoma 2012;53:1155-61. [Crossref] [PubMed]
- Spina V, Rossi D. NF-κB deregulation in splenic marginal zone lymphoma. Semin Cancer Biol 2016;39:61-7. [Crossref] [PubMed]
- Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol 2009;9:767-77. [Crossref] [PubMed]
- Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol 1990;10:2327-34. [Crossref] [PubMed]
- Serfling E, Barthelmäs R, Pfeuffer I, et al. Ubiquitous and lymphocyte-specific factors are involved in the induction of the mouse interleukin 2 gene in T lymphocytes. EMBO J 1989;8:465-73. [Crossref] [PubMed]
- Hendrayani SF, Al-Harbi B, Al-Ansari MM, et al. The inflammatory/cancer-related IL-6/STAT3/NF-κB positive feedback loop includes AUF1 and maintains the active state of breast myofibroblasts. Oncotarget 2016;7:41974-85. [Crossref] [PubMed]
- Thieblemont C, Molina T, Davi F. Optimizing therapy for nodal marginal zone lymphoma. Blood 2016;127:2064-71. [Crossref] [PubMed]
Cite this article as: Spina V, Mensah AA, Arribas AJ. Biology of splenic and nodal marginal zone lymphomas. Ann Lymphoma 2021;5:6.


