
Mucosa-associated lymphoid tissue lymphoma of various sites: common molecular mechanisms but different players
Introduction
Extranodal marginal zone lymphoma of mucosa-associated lymphoid tissue (MALT) lymphoma is a low-grade B-cell lymphoma originated from marginal zone B-cells. The lymphoma commonly arises in a background of a chronic inflammatory disorder at diverse anatomic sites typically devoid of any native lymphoid tissue (1). The cause of the chronic inflammation is highly site dependent, ranging from Helicobacter pylori infection in MALT lymphoma of the stomach to autoimmune responses in those of the salivary gland (Sjogren’s syndrome) and thyroid (Hashimoto’s thyroiditis). The inflammatory response results in development and accumulation of organised lymphoid tissue, i.e., acquired MALT. Like the secondary lymphoid tissue, the acquired MALT generates and maintains adaptive immune responses including T-cell dependent B-cell maturation. Thus, antigen primed B-cells actively undergo the germinal centre reaction, affinity maturation and differentiate into antigen experienced effective B-cells and plasma cells. Through this relentless dynamic process, reactive B-cells may undergo Darwinian selection based on their growth and survival capacity. This may lead to gradual clonal selection, expansion, and eventual malignant transformation of certain reactive B-cell clones. This is supported by the findings of multiple small clonal B-cell populations at reactive conditions, and their occasional progression and clonal expansion to develop an overt lymphoma (2).
In general, MALT lymphoma only develops in a very small number of the patients with these common chronic inflammatory disorders. Apart from the host genetic and microbial virulent factors where indicated, the B-cell clone, which utmost undergoes malignant transformation, is most likely selected owing to its intrinsic properties, appropriate supporting microenvironments and cooperative somatic genetic changes. Although this is not yet fully understood, there is growing evidence indicating oncogenic cooperation among these three distinct components in the clonal evolution of MALT lymphoma. At the molecular level, all the three components affect the cellular signalling pathways critical for the biology of marginal zone B-cells. Nonetheless, there is a distinct difference in the players involved among MALT lymphomas of different sites. This review will focus on the recent advances that advocate for oncogenic cooperation between immunological drive and somatic genetic change in MALT lymphoma of various mucosal sites. To help the understanding of oncogenic role of various players in MALT lymphoma, I first briefly outline below the signalling pathways that are critical for the biology of marginal zone B-cells.
Cellular signalling critical for marginal zone B-cells
Marginal zone B-cells are post-germinal centre B-cells, their differentiation, migration and homing are tightly regulated by concerted actions of transcriptional network and surface receptor signalling. There are several signalling pathways including NF-κB and NOTCH, which are critical for marginal zone B-cells, and disruption of any of these signalling pathways impairs the development and/or function of marginal zone B-cells (3).
NF-κB signalling pathway
Nuclear factor (NF)-κB is a family of dimeric transcription factors, which are formed by various hetero or homodimers of RelA (p65), RelB, c-Rel, NF-κB1 (p50 and its precursor p105) and NF-κB2 (p52 and its precursor p100). They are kept inactive in the cytoplasm by their inhibitor (IκBα, IκBβ and IκBε) or in its dormant precursor form, but can be activated by various surface receptor signalling, enabling their nuclear translocation and transcriptional activities. There are two common pathways linking various surface receptor signalling to NF-κB activation, namely canonical and non-canonical NF-κB pathway.
Canonical NF-κB pathway
This is characterised by activation of the IκB kinase (IKK) complex in response to upstream signalling, which phosphorylates IκB, triggering its K48-linked polyubiquitination and degradation by proteasome. NF-κB dimers are then free for nuclear translocation, enabling their transcriptional activities (Figure 1).
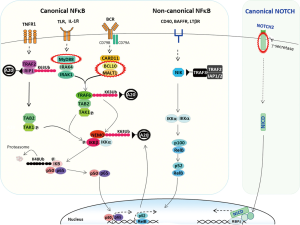
Canonical NF-κB pathway is activated by signalling from several surface receptors including B-cell receptor (BCR), Toll-like receptor (TLR), interleukin 1 receptor (IL1R) and tumour necrosis factor receptor (TNFR). Binding of these receptors by ligand triggers distinct cascade signalling, but converge on the canonical NF-κB activation pathway. For example, BCR engagement triggers phosphorylation of CD79A (Igα) and CD79B (Igβ) at their ITAM (immunoreceptor tyrosine-based activation motif) residues, which act as docking sites for LYN and SYK kinases, and their signal propagation (5). These kinases recruit the B-Cell Linker (BLNK), and coordinate the activation of Bruton's tyrosine kinase (BTK) and phospholipase Cg2 (PLCγ2). In turn, they trigger a signalling cascade that promote the assembly and activation of the CARD11/BCL10/MALT1 (CBM) signalosome complex (6,7), which links the BCR receptor signalling to the canonical NF-κB activation pathway. Apart from the canonical NF-κB pathway, the proximal BCR signalling also activate the PI3K–AKT and RAS-ERK activation pathways via different signalling cascades (5,8,9).
The signalling that leads to NF-κB activation is closely regulated by several negative regulators including TNFα inducible protein 3 (TNFAIP3, also known as A20), IκBα and CYLD (cylindromatosis) (10). Importantly, IκBα and TNFAIP3 are the transcriptional targets of NF-κB, and their expression could act as an auto-negative feedback to ensure appropriate level and length of NF-κB activation. Apart from immobilisation of NF-κB in the cytoplasm, IκBα harbours a nuclear export sequence and can promote NF-κB export from the cell nucleus to the cytoplasm (11). TNFAIP3 is an ubiquitin editing enzyme, can remove the K63-linked ubiquitin chain and catalyse the K48-linked polyubiquitination, which are modifications for protein activation and degradation respectively (12). Through these ubiquitin editing activities, TNFAIP3 can inactivate a number of NF-κB positive regulators including receptor-interacting protein-1/2 (RIP1/2), ubiquitin-conjugating enzyme 13 (Ubc13) and IKKγ (also known as NF-κB essential modulator, NEMO), thus attenuating the signalling from several surface receptors including BCR, TNFR, TLR and IL1βR (13-15).
Non-canonical NF-κB pathway
This is featured by activation of the NF-κB inducible kinase (NIK) in response to upstream signalling. The activated NIK phosphorylates and activates IKKα, which in turn triggers phosphorylation and partial proteolysis of NF-κB2 (p100), generating a functional active form p52 in complex with RelB. The p52/RelB dimer is then free for nuclear translocation and transcriptional function. The non-canonical NF-κB pathway is activated by signalling from surface receptors including CD40, B cell activating factor receptor (BAFFR), TNFRSF13B (also known as TACI) and lymphotoxin β receptor (LTβR). The pathway is modulated by controlling NIK degradation, which is mediated by an ubiquitin ligase complex comprising TRAF3, TRAF2, and cIAP1/2 (API1/2) (16).
NOTCH2 signalling pathway
NOTCH2 is a surface receptor, composed of extracellular, transmembrane and intracellular domain. Binding of its extracellular domain by ligand triggers proteolytic cleavages of NOTCH2 by ADAM-type protease (S2) and γ-secretase (S3), and this releases the active NOTCH2 intracellular domain (N2ICD) (17). N2ICD is free to translocate to the nucleus, forms a transcriptionally active complex with RBPJ (CSL) and transactivate its target genes (17).
The above receptor signalling is important not only for the differentiation, but also for the function of marginal zone B-cell. This is supported by finding of expression of a range of ligands including BAFF, APRIL, CD40L and the Notch2 ligand Delta-like 1 (DLL1) by the innate lymphoid cells that reside in the marginal zone (18,19). These signalling, critical for normal marginal zone B-cells, are also operational in MALT lymphoma cells, and often affected by somatic genetic changes, together causing their constitutive activation.
Receptor signalling commonly implicated in MALT lymphoma of various sites
BCR signalling
Evidence of active BCR signalling in MALT lymphoma
A functional BCR is essential for the biology of B-cells, and this is also true if not more paramount for MALT lymphoma cells. The lymphoma cells invariably express surface IgM, and preserve the biological properties of reactive B-cells such as the ability to undergo blast transformation, plasma cell differentiation as well as the “germinal centre reaction”, histologically known as follicular colonisation (20,21). Similar to germinal centre B-cells, MALT lymphoma cells frequently show intraclonal sequence variations in their rearranged IG genes, a likely consequence of their follicular colonisation (22-26). Importantly, crosslinking surface IgM on MALT lymphoma cells stimulates their proliferative responses to mitogens (27). As expected, inhibiting BCR signalling by BTK inhibitor induces durable responses in patients with previously treated marginal zone lymphoma including MALT lymphoma (28). Together, these observations indicate that BCR signalling is operational in MALT lymphoma cells.
Evidence of auto-reactivity of MALT lymphoma associated BCR
The specificity of immunoglobulin derived from MALT lymphoma has not been investigated in a large scale. Nonetheless, various studies using hybridoma or recombinant antibody technology consistently show that the immunoglobulin from MALT lymphoma of various anatomic sites is autoreactive rather than recognising antigens from infectious agents that are associated with the lymphoma development (Figure 2A). The auto-reactivity may range from polyreactive to various self-antigens to a high-affinity binding to IgG-Fc, a characteristic of rheumatoid factors (RF) (Figure 2). In fact, many of these MALT lymphoma derived immunoglobulins share the cardinal features of known autoantibodies, together providing the paradigm for sequence analysis of lymphoma derived immunoglobulin. This has considerably expanded our knowledge on the extent of autoreactivity of MALT lymphoma associated BCR.
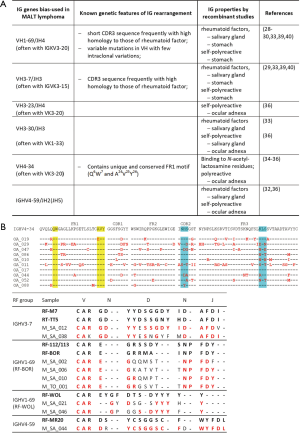
A paradigm is the immunoglobulin displaying RF activities, which are generated commonly by IGHV1-69/J4, IGHV3-7/J3 and rarely by IGHV4-59/J2(J5) rearrangements (33-38). The vast majority of these IG gene rearrangements seen in MALT lymphoma of various anatomic sites show a high identity to those of classical RFs in their complementarity-determining regions (CDR) 3 sequences (Figure 2B). Interestingly, IGHV1-69/J4 is preferentially paired with IGKV3-20 rearrangement, while IGHV3-7/J3 is often paired with IGVK3-15 rearrangement, further emphasizing their selection for antigen binding (38).
Another example is the immunoglobulin encoded by IGHV4-34 rearrangement, which is auto-reactive, binding to the carbohydrate I/i antigens composed of N-acetyl-lactosamine units. The hydrophobic residues (Q6W7A24V25Y26) in the framework region 1 (FR1) of IGHV4-34 are essential for binding to N-acetyl-lactosamine residues, and introduction of mutations in these motifs impair the survival of the lymphoma cells expressing IGHV4-34 BCR (29,30). Remarkably, the germline FR1 hydrophobic patch is spared by somatic mutations in MALT lymphoma despite high levels of somatic mutations in other regions of the IGHV4-34 rearrangement (Figure 2B) (30,32,39).
Evidence of biased IG gene usage in MALT lymphoma
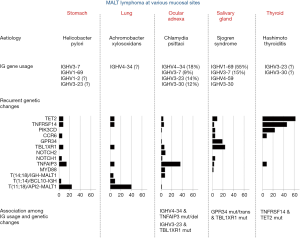
To date, sequencing studies of the rearranged IG genes in MALT lymphoma have been largely focused on IGH in those from the salivary gland, ocular adnexa and stomach. There is a clear evidence for a biased usage of IGH genes in MALT lymphoma of the salivary gland and ocular adnexa (Figure 3). Over 70% of salivary gland MALT lymphomas harbour IGHV1-69/J4 (~55%) or IGHV3-7/J3 (~15%) rearrangements (32,33,35,38). This together with other less encountered (IGHV4-59/J2(J5) and IGHV3-30/JH4) rearrangements indicates that most salivary gland MALT lymphomas express BCR that potentially bears RF activities. Ocular adnexal MALT lymphomas show a biased usage of IGHV4-34 (18%), followed by IGHV3-23 (12–17%), IGHV3-30 (10-14%) and IGHV3–7 (9%) (32,40,41). In contrast, gastric MALT lymphomas generally display more diversified IGH gene usages, but are nonetheless enriched by those coding for autoantibodies. The difference in IG gene usage among MALT lymphoma of different anatomic sites is mostly likely the outcome of adaptive response and clonal selection caused by their distinct aetiologies, hence different antigen exposures (64).
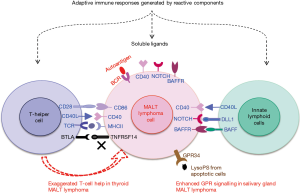
T-cell help and receptor signalling to non-canonical NF-κB pathway
Marginal zone B-cells develop via a T-cell dependent B-cell maturation process. Studies largely based on gastric MALT lymphoma indicate a critical role of T-cell help in the lymphoma development, which may represent a common mechanism for MALT lymphoma as a whole. Early studies show that H. pylori primed tumour infiltrating T-cells stimulate the proliferation of gastric lymphoma B-cells in vitro via cognate interaction involving CD40/CD40L co-stimulating molecules (65-67). These observations are reinforced by animal model studies, showing that depletion of CD4 positive T-cells can block the tumour growth in vivo (68,69). As there is no evidence that the BCR expressed by human gastric MALT lymphoma recognises H. pylori associated antigens, it is unclear how lymphoma B-cells interact with H. pylori specific T-cells without recognising a common antigen. Alternatively, the stimulation of lymphoma B-cells is mediated through bystander T-cell help via soluble ligands and cytokines, such as CD40L and BAFF (Figure 1) (70-72). In support of this speculation, an enriched expression of proinflammatory cytokines such as IL8 and IL1β, molecules involved in B and T-cell interaction such as CD86, CD28 and ICOS is seen in gastric MALT lymphoma, particularly those without chromosome translocation or responsive to H. pylori eradication therapy (73-76). Such bystander T-cell help by H. pylori specific T-cells could be generated by classical immunological responses through reactive B-cell compartments which are always present in gastric MALT lymphoma. Regardless of the mechanistic details, these receptor signalling are critical for the survival of gastric MALT lymphoma cells as 70% of cases show complete lymphoma regression by H. pylori eradication therapy alone (77-79).
Genetic abnormalities
Although there are extensive investigations of genome-wide copy number changes (80,81), there are only three whole exome studies on a small number of MALT lymphoma from the ocular adnexa (n=23), salivary gland (n=14) and thyroid (n=13) (53-55), and several targeted sequencing studies of small gene panels (32,53,54,56). Hence, our understanding of the mutation profile in MALT lymphoma is far from comprehensive. Nonetheless, the data available to date show that there are considerable variations in the incidence and spectrum of genetic abnormalities among MALT lymphoma of different sites (Figure 3). Interestingly, most of these changes affect several common pathways.
Genetic abnormalities that enhance NF-κB signalling
There are several chromosome translocations involving the adaptor molecules that connect the BCR signalling to the NF-κB activation pathway. T(1;14) (p22;q32) and t(14;18) (q32;q21) juxtapose the BCL10 and MALT1 gene under the regulatory control of the IG gene enhancer respectively, causing their over-expression. These translocations are primarily seen in MALT lymphoma of the stomach, lung and ocular adnexa, but at low frequencies (Figure 3) (57). BCL10 and MALT1 over-expression triggers their oligomerisation, hence constitutive activation, leading to enhanced signalling to activate the canonical NF-κB pathway (6,82). MALT1 activation also elicits its protease activities, causing specific cleavage and inactivation of NF-κB negative regulators including TNFAIP3 and CYLD, thus further enhancing NF-κB activation (83-87). BCL10 and MALT1 over-expression may also promote the activation of non-canonical NF-κB pathway via up-regulation of BAFF expression (88,89).
T(11;18)(q21;q21) generates a chimeric fusion between the N-terminal BIRC3 (API2) and the C-terminal MALT1, and is frequently seen in MALT lymphoma of the stomach (24%) and lung (40%) (4,58,90). Through interaction between the BIR1 of the API2 moiety and the C-terminal region of MALT1, the fusion products undergo oligomerisation, become constitutively active, thus triggering signalling to activate the canonical NF-κB pathway (91-93). The API2-MALT1 fusion product can also cleave TNFAIP3 and CYLD through MALT1 protease activities, thus abolishing their negative feedback regulation (83,94). Theoretically, the API2-MALT1 induced NF-κB activation can enhance its own expression since API2 is a transcriptional target of NF-κB (95). Moreover, the API2-MALT1 fusion product gains novel function to activate the non-canonical NF-κB pathway by cleaving NIK at arginine 325, and generating a C-terminal fragment that retains kinase activity and resists to TRAF3 mediated proteasomal degradation (96). Finally, the fusion product can also cleave the tumour suppresser protein LIMA1 (LIM domain and actin-binding protein-1) and generates a novel oncogenic LIM domain-only (LMO) fragment (97). In support of the above observations, MALT lymphomas with BCL10 or MALT1 translocation show strong signature of NF-κB target gene expression (76,98).
Additionally, there are several other signalling molecules, including MYD88, CARD11 and CD79B, which are activated by mutation (32,99-104). Among these, MYD88 mutation, mainly L265P, is relatively frequent, but primarily in ocular adnexal MALT lymphoma, and the mutation enables MYD88 spontaneously assembling an active complex containing IRAK1 and IRAK4, triggering signalling cascade to activate NF-κB, STAT3 and AP1 transcription factors (Figure 1) (104).
Apart from the above positive regulators activated by chromosome translocation or mutation, several negative regulators of the NF-κB signalling pathway are inactivated by mutation and/or deletion, and they include TNFAIP3, TRAF3, TINIP1 and NFKBIA. Among these changes, TNFAIP3 inactivation by mutation and deletion is more frequent, but primarily in MALT lymphoma of the ocular adnexa (36%) and dura (67%) (Figure 1) (59,60,105-109). TNFAIP3 inactivation can enhance the activation of the canonical NF-κB pathway triggered by signalling from several receptors including BCR, TLR and TNFR1 due to loss of negative regulation on several signalling molecules (IKKγ, TRAF6 and RIP1/2) downstream of their receptors (Figure 1) (13,110,111). Similarly, TRAF3 inactivation promotes the activation of the non-canonical NF-κB pathway due to impaired control on NIK degradation (Figure 1).
Genetic changes that enhance NOTCH signalling
Only a few of studies investigated NOTCH signalling pathway for mutation in MALT lymphoma, mainly in those from the ocular adnexa and dura. Nonetheless, these studies revealed recurrent mutations in NOTCH1 and NOTCH2 (56,109,112), and the majority of the observed mutations predict a truncated NOTCH1/2 product lacking the C-terminal PEST domain which regulates the protein stability and degradation. Thus, these mutations are activation changes, most likely enhancing NOTCH stability and prolonging its activity (113).
Genetic abnormalities affecting G protein-coupled receptor (GPR)
The extent of genetic abnormalities in GPR signalling pathways is unclear due to few studies by whole exome and targeted sequencing. Nonetheless, recurrent mutations are seen in GPR34 and CCR6, with GPR34 changes primarily restricted to salivary gland MALT lymphoma (53). The majority of mutations in these GPRs are nonsense changes or frameshift indels, which are clustered in their C-terminal region (53). These mutations predict a truncated GPR product that lacks the C-terminal phosphorylation motif, which is essential for interaction with β-arrestin and receptor desensitization (53). As expected, the GPR34 and CCR6 truncation mutants are more potent in activation of a range of downstream signalling pathways including NF-κB and MAPK/JNK than their wild type counterpart (unpublished data, April 2020). Apart from mutation, GPR34 is also targeted by t(X:14)(p11.4;q32), which cause GPR34 over-expression. Like GPR34 mutation, this translocation is also restricted to MALT lymphoma of salivary gland (114-116).
Genetic abnormalities that deregulate transcriptional control
Recurrent mutations have also been described in several transcriptional regulators including TBL1XR1, TET2 and CREBP, again variably involved in MALT lymphoma of different sites (32,53,54,56,107). TBL1XR1 mutation is frequently seen in MALT lymphoma of the salivary gland (24%), ocular adnexa (6%), while TET2 mutation is biased toward to those of the thyroid (62%). Majority of TBL1XR1 mutations are missense changes affecting regions or residues critical for interaction with NCoR, and may enhance TBL1XR1 binding to NCoR and facilitate its degradation, consequently promoting NF-κB and JUN target gene expression (53,54). While most TET2 mutations are deleterious alterations, likely inactivating or impairing its dioxygenase activity, hence its role in DNA demethylation, consequently affecting a wide spectrum of gene expression (53,117). The biological impact of genetic change in these transcriptional regulators is likely broad, and their oncogenic impact most likely depends on concurrent cooperating events.
Genetic abnormalities that deregulate B and T cell interactions
Despite limited number of MALT lymphomas investigated by whole exome sequencing, studies to date have revealed frequent mutation in TNFRSF14 (HVEM: herpesvirus entry mediator) particularly in thyroid MALT lymphoma (53,107). Majority of TNFRSF14 mutations are deleterious changes such as nonsense, frameshift alterations, thus most likely inactivating or impairing the protein function (53). TNFRSF14 is a surface ligand for T lymphocyte attenuator (BTLA) expressed on T-helper cells, and binding of BTLA by TNFRSF14 attenuates T-cell function, restraining its help to B-cells through downregulation of CD40-CD40L interactions (118). In a mouse model study, Tnfrsf14 deficient B cells show an enhanced growth advantage due to exaggerated T-helper signals (119). Thus, TNFRSF14 inactivation by genetic change may enable malignant B-cells to gain more T-cell help, a molecular mechanism exemplified by early studies in gastric MALT lymphoma (Figure 4).
Distinct oncogenic cooperation among MALT lymphoma of different sites
As discussed above, there are distinct differences in the aetiology and genetic profile among MALT lymphoma of different sites. Different aetiologies clearly trigger disparate adaptive immune responses, which are prone to generate autoreactive B-cells with distinct specificity, and influence their clonal expansion. Different aetiologies also generate distinct microenvironments, which may give rise to different spectrum of genotoxic damages and influence pathogenic event selection, leading to distinct mutation profile seen in MALT lymphoma of various sites. There is mounting evidence showing significant association between certain intrinsic B-cell properties and genetic changes in MALT lymphoma of various sites, suggesting Darwinian selection of these events during the lymphoma cell clonal evolution.
In ocular adnexal MALT lymphoma, there is a significant association between IGHV4-34 usage and TNFAIP3 inactivation, and also between IGHV3-23 usage and TBL1XR1 mutation (32). Both IGHV4-34 and IGHV3-23 rearrangements generate BCR that are likely autoreactive, hence persistent signalling to activate the canonical NF-κB pathway. The chronic BCR signalling is likely further enhanced by loss of downstream negative regulation, i.e., TNFAIP3 inactivation or enhanced degradation of the transcriptional repressor NCoR by mutated TBL1XR1 (54). The acquisition of TNFAIP3 and TBL1XR1 mutation in these autoreactive clones could be the consequence of Darwinian selection for cooperative oncogenic events during their clonal evolution.
In salivary gland MALT lymphoma, the IGHV1-69 usage is mutually exclusive from the genetic changes identified so far, and the genetic event that cooperates with BCR signalling remains to be identified. Nonetheless, there is a significant association between GPR34 mutation/translocation and TBL1XR1 mutation in cases not using the IGHV1-69 gene (53). As discussed above, GPR34 mutation/translocation enhances its receptor signalling, leading to enhanced NF-κB and MAPK/JNK activities, and this could be further enhanced by TBL1XR1 mutations. The ligand for GPR34 is lysophosphatidylserine, which is generated by hydrolysis of membrane phosphatidyserine, and enriched on the surface of apoptotic cells (120). In light of the prominent lymphoepithelial lesions in salivary gland MALT lymphoma and the early appearance of neoplastic B-cells always surrounding these apoptotic cells, it is pertinent to speculate that this is the special niche favouring the survival and expansion of neoplastic cells carrying GPR34 mutations/translocation (Figure 4).
Finally in thyroid MALT lymphoma, there is a significant correlation between TNFRSF14 and TET2 mutation (53). As discussed above, TNFRSF14 inactivation by mutation enables lymphoma B-cells gain more T-cell help as the mutation attenuates the negative regulation by B-cells to T-helper cells (Figure 4) (119). How such enhanced T-cell help, such as CD40 signalling, cooperate with TET2 inactivation is unclear. Nonetheless, TET2 mediated demethylation is critical for expression of lineage specific genes that regulate T-dependent B-cell maturation (117,121,122). Thyroid MALT lymphomas with TET2 mutation show a high level of extensive DNA methylation, particularly in the promoter regions, than those with wild type TET2 (107). Thus, there is an ample scope for oncogenic cooperation between TET2 inactivation and CD40 and other receptor signalling mediated by T-helper cells.
Future prospective
MALT lymphoma is a paradigm for illustration of oncogenic cooperation between adaptive immune responses and acquired somatic genetic changes in the selection, expansion and malignant transformation of the neoplastic clone. Distinct aetiologies and different mutation profiles among MALT lymphoma of various sites provide a dynamic model to explore the interaction between lymphoma cells and their microenvironments during the multi-stages of the lymphoma development. As neither the immmunogenetic nor the somatic genetic profile has been comprehensively investigated in MALT lymphoma of various sites, the lymphoma remains to be a fertile research area for scientific discoveries in the near future. Finally, the recent advances in characterisation of innate lymphoid cells in the marginal zone provide a further avenue to interrogate the interaction between lymphoma cells and their residential microenvironment.
Acknowledgments
Funding: The studies from Professor Ming-Qing Du’s laboratory were supported by research grants from Blood Cancer, U.K., Kay Kendall Leukaemia Fund, Medical Research Council, The Pathological Society of UK and Ireland, and the Addenbrooke’s Charitable Trust.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Francesco Bertoni, Thomas Habermann, Davide Rossi, Emanuele Zucca) for the series “Marginal Zone Lymphomas” published in Annals of Lymphoma. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aol-20-15). The series “Marginal Zone Lymphomas” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Isaacson PG, Du MQ. MALT lymphoma: from morphology to molecules. Nat Rev Cancer 2004;4:644-53. [Crossref] [PubMed]
- Nakamura S, Aoyagi K, Furuse M, et al. B-cell monoclonality precedes the development of gastric MALT lymphoma in Helicobacter pylori-associated chronic gastritis. Am J Pathol 1998;152:1271-9. [PubMed]
- Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol 2009;9:767-77. [Crossref] [PubMed]
- Du MQ. MALT lymphoma: A paradigm of NF-kappaB dysregulation. Semin Cancer Biol 2016;39:49-60. [Crossref] [PubMed]
- Schweighoffer E, Tybulewicz VL. Signalling for B cell survival. Curr Opin Cell Biol 2018;51:8-14. [Crossref] [PubMed]
- Thome M, Weil R. Post-translational modifications regulate distinct functions of CARMA1 and BCL10. Trends Immunol 2007;28:281-8. [Crossref] [PubMed]
- Yang C, David L, Qiao Q, et al. The CBM signalosome: potential therapeutic target for aggressive lymphoma? Cytokine Growth Factor Rev 2014;25:175-83. [Crossref] [PubMed]
- Rickert RC. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol 2013;13:578-91. [Crossref] [PubMed]
- Seda V, Mraz M. B-cell receptor signalling and its crosstalk with other pathways in normal and malignant cells. Eur J Haematol 2015;94:193-205. [Crossref] [PubMed]
- Sun SC, Ley SC. New insights into NF-kappaB regulation and function. Trends Immunol 2008;29:469-78. [Crossref] [PubMed]
- Wuerzberger-Davis SM, Chen Y, Yang DT, et al. Nuclear export of the NF-kappaB inhibitor IkappaBalpha is required for proper B cell and secondary lymphoid tissue formation. Immunity 2011;34:188-200. [Crossref] [PubMed]
- Martens A, Van Loo G. A20 at the Crossroads of Cell Death, Inflammation, and Autoimmunity. Cold Spring Harb Perspect Biol 2020;12:a036418 [Crossref] [PubMed]
- Vereecke L, Beyaert R, Van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol 2009;30:383-91. [Crossref] [PubMed]
- Skaug B, Chen J, Du F, et al. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol Cell 2011;44:559-71. [Crossref] [PubMed]
- Catrysse L, Vereecke L, Beyaert R, et al. A20 in inflammation and autoimmunity. Trends Immunol 2014;35:22-31. [Crossref] [PubMed]
- Yang XD, Sun SC. Targeting signaling factors for degradation, an emerging mechanism for TRAF functions. Immunol Rev 2015;266:56-71. [Crossref] [PubMed]
- Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol 2013;13:427-37. [Crossref] [PubMed]
- Cerutti A, Cols M, Puga I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol 2013;13:118-32. [Crossref] [PubMed]
- Magri G, Miyajima M, Bascones S, et al. Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat Immunol 2014;15:354-64. [Crossref] [PubMed]
- Isaacson PG, Wotherspoon AC, Diss T, et al. Follicular colonization in B-cell lymphoma of mucosa-associated lymphoid tissue. Am J Surg Pathol 1991;15:819-28. [Crossref] [PubMed]
- Isaacson PG, Androulakis Papachristou A, Diss TC, et al. Follicular colonization in thyroid lymphoma. Am J Pathol 1992;141:43-52. [PubMed]
- Du MQ, Xu CF, Diss TC, et al. Intestinal dissemination of gastric mucosa-associated lymphoid tissue lymphoma. Blood 1996;88:4445-51. [Crossref] [PubMed]
- Du M, Diss TC, Xu C, et al. Ongoing mutation in MALT lymphoma immunoglobulin gene suggests that antigen stimulation plays a role in the clonal expansion. Leukemia 1996;10:1190-7. [PubMed]
- Qin Y, Greiner A, Hallas C, et al. Intraclonal offspring expansion of gastric low-grade MALT-type lymphoma: evidence for the role of antigen-driven high-affinity mutation in lymphomagenesis. Lab Invest 1997;76:477-85. [PubMed]
- Thiede C, Alpen B, Morgner A, et al. Ongoing somatic mutations and clonal expansions after cure of Helicobacter pylori infection in gastric mucosa-associated lymphoid tissue B-cell lymphoma. J Clin Oncol 1998;16:3822-31. [Crossref] [PubMed]
- Zhu D, Lossos C, Chapman-Fredricks JR, et al. Biased use of the IGHV4 family and evidence for antigen selection in Chlamydophila psittaci-negative ocular adnexal extranodal marginal zone lymphomas. PLoS One 2011;6:e29114 [Crossref] [PubMed]
- Hussell T, Isaacson PG, Spencer J. Proliferation and differentiation of tumour cells from B-cell lymphoma of mucosa-associated lymphoid tissue in vitro. J Pathol 1993;169:221-7. [Crossref] [PubMed]
- Noy A, de Vos S, Thieblemont C, et al. Targeting Bruton tyrosine kinase with ibrutinib in relapsed/refractory marginal zone lymphoma. Blood 2017;129:2224-32. [Crossref] [PubMed]
- Richardson C, Chida AS, Adlowitz D, et al. Molecular basis of 9G4 B cell autoreactivity in human systemic lupus erythematosus. J Immunol 2013;191:4926-39. [Crossref] [PubMed]
- Young RM, Wu T, Schmitz R, et al. Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc Natl Acad Sci U S A 2015;112:13447-54. [Crossref] [PubMed]
- Małecka A, Troen G, Tierens A, et al. Immunoglobulin heavy and light chain gene features are correlated with primary cold agglutinin disease onset and activity. Haematologica 2016;101:e361-4. [Crossref] [PubMed]
- Moody S, Escudero-Ibarz L, Wang M, et al. Significant association between TNFAIP3 inactivation and biased immunoglobulin heavy chain variable region 4-34 usage in mucosa-associated lymphoid tissue lymphoma. J Pathol 2017;243:3-8. [Crossref] [PubMed]
- Miklos JA, Swerdlow SH, Bahler DW. Salivary gland mucosa-associated lymphoid tissue lymphoma immunoglobulin V(H) genes show frequent use of V1-69 with distinctive CDR3 features. Blood 2000;95:3878-84. [Crossref] [PubMed]
- Bende RJ, Aarts WM, Riedl RG, et al. Among B cell non-Hodgkin's lymphomas, MALT lymphomas express a unique antibody repertoire with frequent rheumatoid factor reactivity. J Exp Med 2005;201:1229-41. [Crossref] [PubMed]
- Hoogeboom R, Bende RJ, van Noesel CJ. MALT lymphoma-derived rheumatoid factors are nonpolyreactive high-affinity antibodies. Blood 2010;116:1818-9. [Crossref] [PubMed]
- Bende RJ, Slot LM, Hoogeboom R, et al. Stereotypic rheumatoid factors that are frequently expressed in mucosa-associated lymphoid tissue-type lymphomas are rare in the labial salivary glands of patients with Sjogren's syndrome. Arthritis Rheumatol 2015;67:1074-83. [Crossref] [PubMed]
- Bende RJ, Janssen J, Wormhoudt TA, et al. Identification of a novel stereotypic IGHV4-59/IGHJ5-encoded B-cell receptor subset expressed by various B-cell lymphomas with high affinity rheumatoid factor activity. Haematologica 2016;101:e200-3. [Crossref] [PubMed]
- Bende RJ, Janssen J, Beentjes A, et al. Salivary gland MALT lymphomas of Sjgren's syndrome patients in majority express rheumatoid factors affinity-selected for IgG. Arthritis Rheumatol 2020; [Epub ahead of print]. [Crossref]
- Zhu D, Bhatt S, Lu X, et al. Chlamydophila psittaci-negative ocular adnexal marginal zone lymphomas express self polyreactive B-cell receptors. Leukemia 2015;29:1587-99. [Crossref] [PubMed]
- van Maldegem F, Wormhoudt TA, Mulder MM, et al. Chlamydia psittaci-negative ocular adnexal marginal zone B-cell lymphomas have biased VH4-34 immunoglobulin gene expression and proliferate in a distinct inflammatory environment. Leukemia 2012;26:1647-53. [Crossref] [PubMed]
- Dagklis A, Ponzoni M, Govi S, et al. Immunoglobulin gene repertoire in ocular adnexal lymphomas: hints on the nature of the antigenic stimulation. Leukemia 2012;26:814-21. [Crossref] [PubMed]
- Bahler DW, Miklos JA, Swerdlow SH. Ongoing Ig gene hypermutation in salivary gland mucosa- associated lymphoid tissue-type lymphomas. Blood 1997;89:3335-44. [Crossref] [PubMed]
- Kurosu K, Yumoto N, Furukawa M, et al. Low-grade pulmonary mucosa-associated lymphoid tissue lymphoma with or without intraclonal variation. Am J Respir Crit Care Med 1998;158:1613-9. [Crossref] [PubMed]
- Coupland SE, Foss HD, Anagnostopoulos I, et al. Immunoglobulin VH gene expression among extranodal marginal zone B-cell lymphomas of the ocular adnexa. Invest Ophthalmol Vis Sci 1999;40:555-62. [PubMed]
- Sato Y, Nakamura N, Nakamura S, et al. Deviated VH4 immunoglobulin gene usage is found among thyroid mucosa-associated lymphoid tissue lymphomas, similar to the usage at other sites, but is not found in thyroid diffuse large B-cell lymphomas. Mod Pathol 2006;19:1578-84. [Crossref] [PubMed]
- Sakuma H, Nakamura T, Uemura N, et al. Immunoglobulin VH gene analysis in gastric MALT lymphomas. Mod Pathol 2007;20:460-6. [Crossref] [PubMed]
- Mannami T, Yoshino T, Oshima K, et al. Clinical, histopathological, and immunogenetic analysis of ocular adnexal lymphoproliferative disorders: characterization of malt lymphoma and reactive lymphoid hyperplasia. Mod Pathol 2001;14:641-9. [Crossref] [PubMed]
- Bahler DW, Kim BK, Gao A, et al. Analysis of immunoglobulin V genes suggests cutaneous marginal zone B-cell lymphomas recognise similar antigens. Br J Haematol 2006;132:571-5. [Crossref] [PubMed]
- Bahler DW, Szankasi P, Kulkarni S, et al. Use of similar immunoglobulin VH gene segments by MALT lymphomas of the ocular adnexa. Mod Pathol 2009;22:833-8. [Crossref] [PubMed]
- Craig VJ, Arnold I, Gerke C, et al. Gastric MALT lymphoma B cells express polyreactive, somatically mutated immunoglobulins. Blood 2010;115:581-91. [Crossref] [PubMed]
- Perez M, Pacchiarotti A, Frontani M, et al. Primary cutaneous B-cell lymphoma is associated with somatically hypermutated immunoglobulin variable genes and frequent use of VH1-69 and VH4-59 segments. Br J Dermatol 2010;162:611-8. [Crossref] [PubMed]
- Michaeli M, Tabibian-Keissar H, Schiby G, et al. Immunoglobulin gene repertoire diversification and selection in the stomach - from gastritis to gastric lymphomas. Front Immunol 2014;5:264. [Crossref] [PubMed]
- Moody S, Thompson JS, Chuang SS, et al. Novel GPR34 and CCR6 mutation and distinct genetic profiles in MALT lymphomas of different sites. Haematologica 2018;103:1329-36. [Crossref] [PubMed]
- Jung H, Yoo HY, Lee SH, et al. The mutational landscape of ocular marginal zone lymphoma identifies frequent alterations in TNFAIP3 followed by mutations in TBL1XR1 and CREBBP. Oncotarget 2017;8:17038-49. [Crossref] [PubMed]
- Johansson P, Klein-Hitpass L, Budeus B, et al. Identifying Genetic Lesions in Ocular Adnexal Extranodal Marginal Zone Lymphomas of the MALT Subtype by Whole Genome, Whole Exome and Targeted Sequencing. Cancers 2020;12:986. [Crossref] [PubMed]
- Johansson P, Klein-Hitpass L, Grabellus F, et al. Recurrent mutations in NF-kappaB pathway components, KMT2D, and NOTCH1/2 in ocular adnexal MALT-type marginal zone lymphomas. Oncotarget 2016;7:62627-39. [Crossref] [PubMed]
- Ye H, Liu H, Attygalle A, et al. Variable frequencies of t(11;18)(q21;q21) in MALT lymphomas of different sites: significant association with CagA strains of H pylori in gastric MALT lymphoma. Blood 2003;102:1012-8. [Crossref] [PubMed]
- Ye H, Gong L, Liu H, et al. MALT lymphoma with t(14;18)(q32;q21)/IGH-MALT1 is characterized by strong cytoplasmic MALT1 and BCL10 expression. J Pathol 2005;205:293-301. [Crossref] [PubMed]
- Chanudet E, Ye H, Ferry J, et al. A20 deletion is associated with copy number gain at the TNFA/B/C locus and occurs preferentially in translocation-negative MALT lymphoma of the ocular adnexa and salivary glands. J Pathol 2009;217:420-30. [Crossref] [PubMed]
- Bi Y, Zeng N, Chanudet E, et al. A20 inactivation in ocular adnexal MALT lymphoma. Haematologica 2012;97:926-30. [Crossref] [PubMed]
- Liu H, Ye H, Ruskone-Fourmestraux A, et al. T(11;18) is a marker for all stage gastric MALT lymphomas that will not respond to H. pylori eradication. Gastroenterology 2002;122:1286-94. [Crossref] [PubMed]
- Goatly A, Bacon CM, Nakamura S, et al. FOXP1 abnormalities in lymphoma: translocation breakpoint mapping reveals insights into deregulated transcriptional control. Mod Pathol 2008;21:902-11. [Crossref] [PubMed]
- Chanudet E, Huang Y, Ichimura K, et al. A20 is targeted by promoter methylation, deletion and inactivating mutation in MALT lymphoma. Leukemia 2010;24:483-7. [Crossref] [PubMed]
- Xochelli A, Bikos V, Polychronidou E, et al. Disease-biased and shared characteristics of the immunoglobulin gene repertoires in marginal zone B cell lymphoproliferations. J Pathol 2019;247:416-21. [Crossref] [PubMed]
- Hussell T, Isaacson PG, Crabtree JE, et al. Immunoglobulin specificity of low grade B cell gastrointestinal lymphoma of mucosa-associated lymphoid tissue (MALT) type. Am J Pathol 1993;142:285-92. [PubMed]
- Greiner A, Knorr C, Qin Y, et al. CD40 ligand and autoantigen are involved in the pathogenesis of low-grade B-cell lymphomas of mucosa-associated lymphoid tissue. Dev Immunol 1998;6:187-95. [Crossref] [PubMed]
- D'Elios MM, Amedei A, Manghetti M, et al. Impaired T-cell regulation of B-cell growth in Helicobacter pylori--related gastric low-grade MALT lymphoma. GASTROENTEROLOGY 1999;117:1105-12. [Crossref] [PubMed]
- Craig VJ, Cogliatti SB, Arnold I, et al. B-cell receptor signaling and CD40 ligand-independent T cell help cooperate in Helicobacter-induced MALT lymphomagenesis. Leukemia 2010;24:1186-96. [Crossref] [PubMed]
- Hömig-Hölzel C, Hojer C, Rastelli J, et al. Constitutive CD40 signaling in B cells selectively activates the noncanonical NF-kappaB pathway and promotes lymphomagenesis. J Exp Med 2008;205:1317-29. [Crossref] [PubMed]
- Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science 2002;298:2199-202. [Crossref] [PubMed]
- Rodig SJ, Shahsafaei A, Li B, et al. BAFF-R, the major B cell-activating factor receptor, is expressed on most mature B cells and B-cell lymphoproliferative disorders. Hum Pathol 2005;36:1113-9. [Crossref] [PubMed]
- Kuo SH, Tsai HJ, Lin CW, et al. The B-cell-activating factor signalling pathway is associated with Helicobacter pylori independence in gastric mucosa-associated lymphoid tissue lymphoma without t(11;18)(q21;q21). J Pathol 2017;241:420-33. [Crossref] [PubMed]
- de Jong D, Vyth-Dreese F, Dellemijn T, et al. Histological and immunological parameters to predict treatment outcome of Helicobacter pylori eradication in low-grade gastric MALT lymphoma. J Pathol 2001;193:318-24. [Crossref] [PubMed]
- Kuo SH, Chen LT, Chen CL, et al. Expression of CD86 and increased infiltration of NK cells are associated with Helicobacter pylori-dependent state of early stage high-grade gastric MALT lymphoma. World J Gastroenterol 2005;11:4357-62. [Crossref] [PubMed]
- Mueller A, O'Rourke J, Chu P, et al. The role of antigenic drive and tumor-infiltrating accessory cells in the pathogenesis of helicobacter-induced mucosa-associated lymphoid tissue lymphoma. Am J Pathol 2005;167:797-812. [Crossref] [PubMed]
- Hamoudi RA, Appert A, Ye H, et al. Differential expression of NF-kappaB target genes in MALT lymphoma with and without chromosome translocation: insights into molecular mechanism. Leukemia 2010;24:1487-97. [Crossref] [PubMed]
- Du MQ, Atherton JC. Molecular subtyping of gastric MALT lymphomas: implications for prognosis and management. Gut 2006;55:886-93. [Crossref] [PubMed]
- Nakamura S, Ye H, Bacon CM, et al. Clinical impact of genetic aberrations in gastric MALT lymphoma: a comprehensive analysis using interphase fluorescence in situ hybridisation. Gut 2007;56:1358-63. [Crossref] [PubMed]
- Ruskoné-Fourmestraux A, Fischbach W, Aleman BM, et al. EGILS consensus report. Gastric extranodal marginal zone B-cell lymphoma of MALT. Gut 2011;60:747-58. [Crossref] [PubMed]
- Kwee I, Rancoita PMV, Rinaldi A, et al. Genomic profiles of MALT lymphomas: variability across anatomical sites. Haematologica 2011;96:1064-6. [Crossref] [PubMed]
- Rinaldi A, Mian M, Chigrinova E, et al. Blood 2011;117:1595-604. [Crossref] [PubMed]
- Turvey SE, Durandy A, Fischer A, et al. The CARD11-BCL10-MALT1 (CBM) signalosome complex: Stepping into the limelight of human primary immunodeficiency. J Allergy Clin Immunol 2014;134:276-84. [Crossref] [PubMed]
- Coornaert B, Baens M, Heyninck K, et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol 2008;9:263-71. [Crossref] [PubMed]
- Düwel M, Welteke V, Oeckinghaus A, et al. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol 2009;182:7718-28. [Crossref] [PubMed]
- Kirchhofer D, Vucic D. Protease activity of MALT1: a mystery unravelled. Biochem J 2012;444:e3-5. [Crossref] [PubMed]
- Bartuzi P, Hofker MH. van de SB. Tuning NF-kappaB activity: a touch of COMMD proteins. Biochim Biophys Acta 2013;1832:2315-21. [Crossref] [PubMed]
- Hailfinger S, Nogai H, Pelzer C, et al. Malt1-dependent RelB cleavage promotes canonical NF-kappaB activation in lymphocytes and lymphoma cell lines. Proc Natl Acad Sci U S A 2011;108:14596-601. [Crossref] [PubMed]
- Li Z, Wang H, Xue L, et al. Emu-BCL10 mice exhibit constitutive activation of both canonical and noncanonical NF-kappaB pathways generating marginal zone (MZ) B-cell expansion as a precursor to splenic MZ lymphoma. Blood 2009;114:4158-68. [Crossref] [PubMed]
- Tusche MW, Ward LA, Vu F, et al. Differential requirement of MALT1 for BAFF-induced outcomes in B cell subsets. J Exp Med 2009;206:2671-83. [Crossref] [PubMed]
- Streubel B, Simonitsch-Klupp I, Mullauer L, et al. Variable frequencies of MALT lymphoma-associated genetic aberrations in MALT lymphomas of different sites. Leukemia 2004;18:1722-6. [Crossref] [PubMed]
- Lucas PC, Kuffa P, Gu S, et al. A dual role for the API2 moiety in API2-MALT1-dependent NF-kappaB activation: heterotypic oligomerization and TRAF2 recruitment. Oncogene 2007;26:5643-54. [Crossref] [PubMed]
- Zhou H, Du MQ, Dixit VM. Constitutive NF-kappaB activation by the t(11;18)(q21;q21) product in MALT lymphoma is linked to deregulated ubiquitin ligase activity. Cancer Cell 2005;7:425-31. [Crossref] [PubMed]
- Baens M, Fevery S, Sagaert X, et al. Selective expansion of marginal zone B cells in Emicro-API2-MALT1 mice is linked to enhanced IkappaB kinase gamma polyubiquitination. Cancer Res 2006;66:5270-7. [Crossref] [PubMed]
- Staal J, Driege Y, Bekaert T, et al. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J 2011;30:1742-52. [Crossref] [PubMed]
- Hosokawa Y, Suzuki H, Nakagawa M, et al. API2-MALT1 fusion protein induces transcriptional activation of the API2 gene through NF-kappaB binding elements: evidence for a positive feed-back loop pathway resulting in unremitting NF-kappaB activation. Biochem Biophys Res Commun 2005;334:51-60. [Crossref] [PubMed]
- Rosebeck S, Madden L, Jin X, et al. Cleavage of NIK by the API2-MALT1 fusion oncoprotein leads to noncanonical NF-kappaB activation. Science 2011;331:468-72. [Crossref] [PubMed]
- Nie Z, Du MQ, McAllister-Lucas LM, et al. Conversion of the LIMA1 tumour suppressor into an oncogenic LMO-like protein by API2-MALT1 in MALT lymphoma. Nat Commun 2015;6:5908. [Crossref] [PubMed]
- Chng WJ, Remstein ED, Fonseca R, et al. Gene expression profiling of pulmonary mucosa-associated lymphoid tissue lymphoma identifies new biologic insights with potential diagnostic and therapeutic applications. Blood 2009;113:635-45. [Crossref] [PubMed]
- Li ZM, Rinaldi A, Cavalli A, et al. MYD88 somatic mutations in MALT lymphomas. Br J Haematol 2012;158:662-4. [Crossref] [PubMed]
- Liu F, Karube K, Kato H, et al. Mutation analysis of NF-kappaB signal pathway-related genes in ocular MALT lymphoma. Int J Clin Exp Pathol 2012;5:436-41. [PubMed]
- Yan Q, Wang M, Moody S, et al. Distinct involvement of NF-kappaB regulators by somatic mutation in ocular adnexal malt lymphoma. Br J Haematol 2013;160:851-4. [Crossref] [PubMed]
- Gachard N, Parrens M, Soubeyran I, et al. IGHV gene features and MYD88 L265P mutation separate the three marginal zone lymphoma entities and Waldenstrom macroglobulinemia/lymphoplasmacytic lymphomas. Leukemia 2013;27:183-9. [Crossref] [PubMed]
- Zhu D, Ikpatt OF, Dubovy SR, et al. Molecular and genomic aberrations in Chlamydophila psittaci negative ocular adnexal marginal zone lymphomas. Am J Hematol 2013;88:730-5. [Crossref] [PubMed]
- Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011;470:115-9. [Crossref] [PubMed]
- Honma K, Tsuzuki S, Nakagawa M, et al. TNFAIP3 is the target gene of chromosome band 6q23.3-q24.1 loss in ocular adnexal marginal zone B cell lymphoma. Genes Chromosomes Cancer 2008;47:1-7. [Crossref] [PubMed]
- Hyeon J, Lee B, Shin SH, et al. Targeted deep sequencing of gastric marginal zone lymphoma identified alterations of TRAF3 and TNFAIP3 that were mutually exclusive for MALT1 rearrangement. Mod Pathol 2018;31:1418-28. [Crossref] [PubMed]
- Cascione L, Rinaldi A, Bruscaggin A, et al. Novel insights into the genetics and epigenetics of MALT lymphoma unveiled by next generation sequencing analyses. Haematologica 2019;104:e558-61. [Crossref] [PubMed]
- Vela V, Juskevicius D, Gerlach MM, et al. High throughput sequencing reveals high specificity of TNFAIP3 mutations in ocular adnexal marginal zone B-cell lymphomas. Hematol Oncol 2020;38:284-92. [Crossref] [PubMed]
- Ganapathi KA, Jobanputra V, Iwamoto F, et al. The genetic landscape of dural marginal zone lymphomas. Oncotarget 2016;7:43052-61. [Crossref] [PubMed]
- Boone DL, Turer EE, Lee EG, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 2004;5:1052-60. [Crossref] [PubMed]
- Thome M, Tschopp J. TCR-induced NF-kappaB activation: a crucial role for Carma1, Bcl10 and MALT1. Trends Immunol 2003;24:419-24. [Crossref] [PubMed]
- Jangam D, Sridhar K, Butzmann A, et al. TBL1XR1 mutations in primary marginal zone lymphomas of ocular adnexa are associated with unique morphometric phenotypes. Curr Eye Res 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Baumgärtner AK, Zettl A, Chott A, et al. High frequency of genetic aberrations in enteropathy-type T-cell lymphoma. Lab Invest 2003;83:1509-16. [Crossref] [PubMed]
- Baens M, Finalet FJ, Tousseyn T, et al. t(X;14)(p11.4;q32.33) is recurrent in marginal zone lymphoma and up-regulates GPR34. Haematologica 2012;97:184-8. [Crossref] [PubMed]
- Ansell SM, Akasaka T, McPhail E, et al. t(X;14)(p11;q32) in MALT lymphoma involving GPR34 reveals a role for GPR34 in tumor cell growth. Blood 2012;120:3949-57. [Crossref] [PubMed]
- Akasaka T, Lee YF, Novak AJ, et al. Clinical, histopathological, and molecular features of mucosa-associated lymphoid tissue (MALT) lymphoma carrying the t(X;14) (p11;q32)/GPR34-immunoglobulin heavy chain gene. Leuk Lymphoma 2017;58:1-4. [Crossref] [PubMed]
- Shingleton JR, Dave SS. TET2 Deficiency Sets the Stage for B-cell Lymphoma. Cancer Discov 2018;8:1515-7. [Crossref] [PubMed]
- Kennedy R, Klein U A T. Cell-B Cell Tumor-Suppressive Axis in the Germinal Center. Immunity 2019;51:204-6. [Crossref] [PubMed]
- Mintz MA, Felce JH, Chou MY, et al. The HVEM-BTLA Axis Restrains T Cell Help to Germinal Center B Cells and Functions as a Cell-Extrinsic Suppressor in Lymphomagenesis. Immunity 2019;51:310-23.e7. [Crossref] [PubMed]
- Makide K, Aoki J. GPR34 as a lysophosphatidylserine receptor. J Biochem 2013;153:327-9. [Crossref] [PubMed]
- Dominguez PM, Ghamlouch H, Rosikiewicz W, et al. TET2 Deficiency Causes Germinal Center Hyperplasia, Impairs Plasma Cell Differentiation, and Promotes B-cell Lymphomagenesis. Cancer Discov 2018;8:1632-53. [PubMed]
- Schoeler K, Aufschnaiter A, Messner S, et al. TET enzymes control antibody production and shape the mutational landscape in germinal centre B cells. FEBS J 2019;286:3566-81. [Crossref] [PubMed]
Cite this article as: Du MQ. Mucosa-associated lymphoid tissue lymphoma of various sites: common molecular mechanisms but different players. Ann Lymphoma 2020;4:8.


